Insights from genomic profiling of transcription factors
- PMID: 19668247
- PMCID: PMC2846386
- DOI: 10.1038/nrg2636
Insights from genomic profiling of transcription factors
Abstract
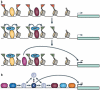
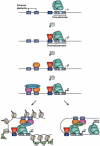
A crucial question in the field of gene regulation is whether the location at which a transcription factor binds influences its effectiveness or the mechanism by which it regulates transcription. Comprehensive transcription factor binding maps are needed to address these issues, and genome-wide mapping is now possible thanks to the technological advances of ChIP-chip and ChIP-seq. This Review discusses how recent genomic profiling of transcription factors gives insight into how binding specificity is achieved and what features of chromatin influence the ability of transcription factors to interact with the genome. It also suggests future experiments that may further our understanding of the causes and consequences of transcription factor-genome interactions.
Figures
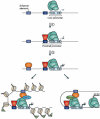
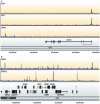
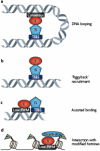
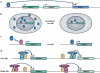
References
-
-
Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 2000;34:77–137. A detailed review of transcriptional regulation, general factors, and accessory proteins that control transcription initation and elongation.
-
-
- Sandelin A, et al. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Reviews Genetics. 2007;8:424–436. - PubMed
-
-
Consortium TEP. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. Demonstrates how genome-wide studies of transcription, factor binding, chromatin structure, DNA replication, and sequence conservation can synergize.
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

