Phenotypic screen of early-developing larvae of the blood fluke, schistosoma mansoni, using RNA interference
- PMID: 19668375
- PMCID: PMC2719580
- DOI: 10.1371/journal.pntd.0000502
Phenotypic screen of early-developing larvae of the blood fluke, schistosoma mansoni, using RNA interference
Erratum in
- PLoS Negl Trop Dis. 2009;3(9):10. doi: 10.1371/annotation/ffd29b50-4102-46b9-b6a1-cdb4984df278.. de Moraes Mourão, Marina [corrected to Mourão, M M].
Abstract
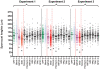
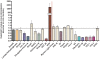
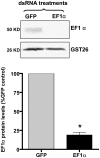
RNA interference (RNAi) represents the only method currently available for manipulating gene-specific expression in Schistosoma spp., although application of this technology as a functional genomic profiling tool has yet to be explored. In the present study 32 genes, including antioxidants, transcription factors, cell signaling molecules and metabolic enzymes, were selected to determine if gene knockdown by RNAi was associated with morphologically definable phenotypic changes in early intramolluscan larval development. Transcript selection was based on their high expression in in vitro cultured S. mansoni primary sporocysts and/or their potential involvement in developmental processes. Miracidia were allowed to transform to sporocysts in the presence of synthesized double-stranded RNAs (dsRNAs) and cultivated for 7 days, during which time developing larvae were closely observed for phenotypic changes including failure/delay in transformation, loss of motility, altered growth and death. Of the phenotypes evaluated, only one was consistently detected; namely a reduction in sporocyst size based on length measurements. The size-reducing phenotype was observed in 11 of the 33 (33%) dsRNA treatment groups, and of these 11 phenotype-associated genes (superoxide dismutase, Smad1, RHO2, Smad2, Cav2A, ring box, GST26, calcineurin B, Smad4, lactate dehydrogenase and EF1alpha), only 6 demonstrated a significant and consistent knockdown of specific transcript expression. Unexpectedly one phenotype-linked gene, superoxide dismutase (SOD), was highly induced ( approximately 1600-fold) upon dsRNA exposure. Variation in dsRNA-mediated silencing effects also was evident in the group of sporocysts that lacked any definable phenotype. Out of 22 nonphenotype-expressing dsRNA treatments (myosin, PKCB, HEXBP, calcium channel, Sma2, RHO1, PKC receptor, DHHC, PepcK, calreticulin, calpain, Smeg, 14.3.3, K5, SPO1, SmZF1, fibrillarin, GST28, GPx, TPx1, TPx2 and TPx2/TPx1), 12 were assessed for the transcript levels. Of those, 6 genes exhibited consistent reductions in steady-state transcript levels, while expression level for the rest remained unchanged. Results demonstrate that the efficacy of dsRNA-treatment in producing consistent phenotypic changes and/or altered gene expression levels in S. mansoni sporocysts is highly dependent on the selected gene (or the specific dsRNA sequence used) and the timing of evaluation after treatment. Although RNAi holds great promise as a functional genomics tool for larval schistosomes, our finding of potential off-target or nonspecific effects of some dsRNA treatments and variable efficiencies in specific gene knockdown indicate a critical need for gene-specific testing and optimization as an essential part of experimental design, execution and data interpretation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Role of the endogenous antioxidant system in the protection of Schistosoma mansoni primary sporocysts against exogenous oxidative stress.PLoS Negl Trop Dis. 2009 Nov 17;3(11):e550. doi: 10.1371/journal.pntd.0000550. PLoS Negl Trop Dis. 2009. PMID: 19924224 Free PMC article.
-
Using RNA interference to manipulate endogenous gene expression in Schistosoma mansoni sporocysts.Mol Biochem Parasitol. 2003 May;128(2):205-15. doi: 10.1016/s0166-6851(03)00078-1. Mol Biochem Parasitol. 2003. PMID: 12742587
-
Potential role of a CD36-like class B scavenger receptor in the binding of modified low-density lipoprotein (acLDL) to the tegumental surface of Schistosoma mansoni sporocysts.Mol Biochem Parasitol. 2006 Apr;146(2):219-30. doi: 10.1016/j.molbiopara.2005.12.010. Epub 2006 Jan 6. Mol Biochem Parasitol. 2006. PMID: 16427708
-
Calmodulin disruption impacts growth and motility in juvenile liver fluke.Parasit Vectors. 2016 Jan 27;9:46. doi: 10.1186/s13071-016-1324-9. Parasit Vectors. 2016. PMID: 26817678 Free PMC article.
-
RNA interference in parasitic helminths: current situation, potential pitfalls and future prospects.Parasitology. 2007 May;134(Pt 5):609-19. doi: 10.1017/S0031182006002071. Epub 2007 Jan 4. Parasitology. 2007. PMID: 17201997 Review.
Cited by
-
Echinococcus granulosus sensu stricto: silencing of thioredoxin peroxidase impairs the differentiation of protoscoleces into metacestodes.Parasite. 2018;25:57. doi: 10.1051/parasite/2018055. Epub 2018 Nov 26. Parasite. 2018. PMID: 30474598 Free PMC article.
-
Calmodulin-specific small interfering RNA induces consistent expression suppression and morphological changes in Echinococcus granulosus.Sci Rep. 2019 Mar 7;9(1):3894. doi: 10.1038/s41598-019-40656-w. Sci Rep. 2019. PMID: 30846822 Free PMC article.
-
Genetic manipulation of schistosomes--progress with integration competent vectors.Parasitology. 2012 Apr;139(5):641-50. doi: 10.1017/S003118201100134X. Epub 2011 Aug 19. Parasitology. 2012. PMID: 21854676 Free PMC article. Review.
-
Tuning of peroxiredoxin catalysis for various physiological roles.Biochemistry. 2014 Dec 16;53(49):7693-705. doi: 10.1021/bi5013222. Epub 2014 Dec 1. Biochemistry. 2014. PMID: 25403613 Free PMC article. Review.
-
Curcumin Generates Oxidative Stress and Induces Apoptosis in Adult Schistosoma mansoni Worms.PLoS One. 2016 Nov 22;11(11):e0167135. doi: 10.1371/journal.pone.0167135. eCollection 2016. PLoS One. 2016. PMID: 27875592 Free PMC article.
References
-
- http://www.who.int/tdr/publications/tdr-research-publications/swg-report... Scientific working group on Schistosomiasis Meeting report. 14–16 November 2005,Geneva, Switzerland.
-
- Steinmann P, Keiser J, Bos R, Tanner M, Utzinger J. Schistosomiasis and water resources development: systematic review, meta-analysis, and estimates of people at risk. Lancet Infect Dis. 2006;6:411–425. - PubMed
-
- van der Werf MJ, de Vlas SJ, Brooker S, Looman CW, Nagelkerke NJ, et al. Quantification of clinical morbidity associated with schistosome infection in sub-Saharan Africa. Acta Trop. 2003;86:125–139. - PubMed
-
- Hotez P, Ottesen E, Fenwick A, Molyneux D. The neglected tropical diseases: the ancient afflictions of stigma and poverty and the prospects for their control and elimination. Adv Exp Med Biol. 2006;582:23–33. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

