HDF1 and RAD17 genes are involved in DNA double-strand break repair in stationary phase Saccharomyces cerevisiae
- PMID: 19669493
- PMCID: PMC2577742
- DOI: 10.1007/s10867-008-9105-0
HDF1 and RAD17 genes are involved in DNA double-strand break repair in stationary phase Saccharomyces cerevisiae
Abstract
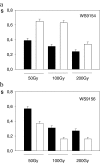
DNA repair, checkpoint pathways and protection mechanisms against different types of perturbations are critical factors for the prevention of genomic instability. The aim of the present work was to analyze the roles of RAD17 and HDF1 gene products during the late stationary phase, in haploid and diploid yeast cells upon gamma irradiation. The checkpoint protein, Rad17, is a component of a PCNA-like complex-the Rad17/Mec3/Ddc1 clamp-acting as a damage sensor; this protein is also involved in double-strand break (DBS) repair in cycling cells. The HDF1 gene product is a key component of the non-homologous end-joining pathway (NHEJ). Diploid and haploid rad17Delta/rad17Delta, and hdf1Delta Saccharomyces cerevisiae mutant strains and corresponding isogenic wild types were used in the present study. Yeast cells were grown in standard liquid nutrient medium, and maintained at 30 degrees C for 21 days in the stationary phase, without added nutrients. Cell samples were irradiated with (60)Co gamma rays at 5 Gy/s, 50 Gy <or= Dabs <or= 200 Gy. Thereafter, cells were incubated in PBS (liquid holding: LH, 0 <or= t <or= 24 h). DNA chromosomal analysis (by pulsed-field electrophoresis), and surviving fractions were determined as a function of absorbed doses, either immediately after irradiation or after LH. Our results demonstrated that the proteins Rad17, as well as Hdf1, play essential roles in DBS repair and survival after gamma irradiation in the late stationary phase and upon nutrient stress (LH after irradiation). In haploid cells, the main pathway is NHEJ. In the diploid state, the induction of LH recovery requires the function of Rad17. Results are compatible with the action of a network of DBS repair pathways expressed upon different ploidies, and different magnitudes of DNA damage.
Figures




Similar articles
-
Roles of Saccharomyces cerevisiae RAD17 and CHK1 checkpoint genes in the repair of double-strand breaks in cycling cells.Radiat Environ Biophys. 2007 Nov;46(4):401-7. doi: 10.1007/s00411-007-0119-y. Epub 2007 Jul 12. Radiat Environ Biophys. 2007. PMID: 17624540
-
Abasic sites linked to dUTP incorporation in DNA are a major cause of spontaneous mutations in absence of base excision repair and Rad17-Mec3-Ddc1 (9-1-1) DNA damage checkpoint clamp in Saccharomyces cerevisiae.DNA Repair (Amst). 2012 Mar 1;11(3):294-303. doi: 10.1016/j.dnarep.2011.12.004. Epub 2012 Jan 4. DNA Repair (Amst). 2012. PMID: 22226374
-
Role of a complex containing Rad17, Mec3, and Ddc1 in the yeast DNA damage checkpoint pathway.Mol Cell Biol. 1999 Feb;19(2):1136-43. doi: 10.1128/MCB.19.2.1136. Mol Cell Biol. 1999. PMID: 9891048 Free PMC article.
-
Mechanisms of DNA double strand break repair and chromosome aberration formation.Cytogenet Genome Res. 2004;104(1-4):14-20. doi: 10.1159/000077461. Cytogenet Genome Res. 2004. PMID: 15162010 Review.
-
[Double strand break repair, one mechanism can hide another: alternative non-homologous end joining].Cancer Radiother. 2012 Feb;16(1):1-10. doi: 10.1016/j.canrad.2011.05.004. Epub 2011 Jul 6. Cancer Radiother. 2012. PMID: 21737335 Review. French.
Cited by
-
The transcription factor, Nuclear factor, erythroid 2 (Nfe2), is a regulator of the oxidative stress response during Danio rerio development.Aquat Toxicol. 2016 Nov;180:141-154. doi: 10.1016/j.aquatox.2016.09.019. Epub 2016 Oct 1. Aquat Toxicol. 2016. PMID: 27716579 Free PMC article.
-
The non-homologous end-joining pathway of S. cerevisiae works effectively in G1-phase cells, and religates cognate ends correctly and non-randomly.DNA Repair (Amst). 2016 Jun;42:1-10. doi: 10.1016/j.dnarep.2016.03.013. Epub 2016 Apr 14. DNA Repair (Amst). 2016. PMID: 27130982 Free PMC article.
-
Analysis of radioprotection and antimutagenic effects of Ilex paraguariensis infusion and its component rutin.Braz J Med Biol Res. 2018 Jul 16;51(9):e7404. doi: 10.1590/1414-431X20187404. Braz J Med Biol Res. 2018. PMID: 30020319 Free PMC article.
References
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1126/science.2683079', 'is_inner': False, 'url': 'https://doi.org/10.1126/science.2683079'}, {'type': 'PubMed', 'value': '2683079', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/2683079/'}]}
- Hartwell, L., Weinert, T.A.: Checkpoints: controls that ensure the order of cell cycle events. Science 246, 629–634 (1989). doi:10.1126/science.2683079 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1126/science.274.5293.1664', 'is_inner': False, 'url': 'https://doi.org/10.1126/science.274.5293.1664'}, {'type': 'PubMed', 'value': '8939848', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/8939848/'}]}
- Elledge, S.J.: Cell cycle checkpoints: preventing an identity crisis. Science 274, 1664–1672 (1996). doi:10.1126/science.274.5293.1664 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1146/annurev.genet.36.060402.113540', 'is_inner': False, 'url': 'https://doi.org/10.1146/annurev.genet.36.060402.113540'}, {'type': 'PubMed', 'value': '12429704', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/12429704/'}]}
- Nyberg, K.A., Michelson, R.J., Putnam, C.W., Weinert, T.A.: Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36, 617–656 (2002). doi:10.1146/annurev.genet.36.060402.113540 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'PubMed', 'value': '11356855', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/11356855/'}]}
- Zhang, H., Zhu, Z., Vidanes, G., Mbangkollo, D., Liu, Y., Siede, W.: Characterization of DNA damage-stimulated self-interaction of Saccharomyces cerevisiae checkpoint protein Rad17p. J. Biol. Chem. 276, 26715–16723 (2001). doi:10.1074/jbc.M103682200 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1073/pnas.0437148100', 'is_inner': False, 'url': 'https://doi.org/10.1073/pnas.0437148100'}, {'type': 'PMC', 'value': 'PMC151326', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC151326/'}, {'type': 'PubMed', 'value': '12604797', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/12604797/'}]}
- Majka, J., Burgers, P.M.: Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc. Natl. Acad. Sci. U.S.A. 100, 2249–2254 (2003). doi:10.1073/pnas.0437148100 - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

