Regulation of the F1F0-ATP synthase rotary nanomotor in its monomeric-bacterial and dimeric-mitochondrial forms
- PMID: 19669503
- PMCID: PMC2577739
- DOI: 10.1007/s10867-008-9114-z
Regulation of the F1F0-ATP synthase rotary nanomotor in its monomeric-bacterial and dimeric-mitochondrial forms
Abstract
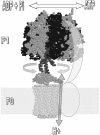
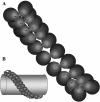
The F(1)F(0)-adenosine triphosphate (ATP) synthase rotational motor synthesizes most of the ATP required for living from adenosine diphosphate, Pi, and a proton electrochemical gradient across energy-transducing membranes of bacteria, chloroplasts, and mitochondria. However, as a reversible nanomotor, it also hydrolyzes ATP during de-energized conditions in all energy-transducing systems. Thus, different subunits and mechanisms have emerged in nature to control the intrinsic rotation of the enzyme to favor the ATP synthase activity over its opposite and commonly wasteful ATPase turnover. Recent advances in the structural analysis of the bacterial and mitochondrial ATP synthases are summarized to review the distribution and mechanism of the subunits that are part of the central rotor and regulate its gyration. In eubacteria, the epsilon subunit works as a ratchet to favor the rotation of the central stalk in the ATP synthase direction by extending and contracting two alpha-helixes of its C-terminal side and also by binding ATP with low affinity in thermophilic bacteria. On the other hand, in bovine heart mitochondria, the so-called inhibitor protein (IF(1)) interferes with the intrinsic rotational mechanism of the central gamma subunit and with the opening and closing of the catalytic beta-subunits to inhibit its ATPase activity. Besides its inhibitory role, the IF(1) protein also promotes the dimerization of the bovine and rat mitochondrial enzymes, albeit it is not essential for dimerization of the yeast F(1)F(0) mitochondrial complex. High-resolution electron microscopy of the dimeric enzyme in its bovine and yeast forms shows a conical shape that is compatible with the role of the ATP synthase dimer in the formation of tubular the cristae membrane of mitochondria after further oligomerization. Dimerization of the mitochondrial ATP synthase diminishes the rotational drag of the central rotor that would decrease the coupling efficiency between rotation of the central stalk and ATP synthesis taking place at the F(1) portion. In addition, F(1)F(0) dimerization and its further oligomerization also increase the stability of the enzyme to natural or experimentally induced destabilizing conditions.
Figures



References
-
- Penefsky, H.S., Pullman, M.E., Datta, A., Racker, E.: Partial resolution of the enzymes catalyzing oxidative phosphorylation. II. Participation of a soluble adenosine triphosphatase in oxidative phosphorylation. J. Biol. Chem. 235, 3330–3336 (1960) - PubMed
-
- García, J.J., Gómez-Puyou, A., Maldonado, E., Tuena de Gómez-Puyou, M.: Acceleration of unisite catalysis of mitochondrial F1-adenosinetriphosphatase by ATP, ADP and pyrophosphate—hydrolysis and release of the previously bound [gamma-32P]ATP. Eur. J. Biochem. 249(2), 622–229 (1997). doi:10.1111/j.1432-1033.1997.00622.x - DOI - PubMed
-
- García, J.J.: The F0F1-ATP synthase: binding energy, coupling and rotational catalysis. In: Pandalai, S.G. (ed.) Recent Research Developments in Bioenergetics, p. 41. Transworld Research Network, Trivandrum (2000)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

