Large Arf1 guanine nucleotide exchange factors: evolution, domain structure, and roles in membrane trafficking and human disease
- PMID: 19669794
- PMCID: PMC7088145
- DOI: 10.1007/s00438-009-0473-3
Large Arf1 guanine nucleotide exchange factors: evolution, domain structure, and roles in membrane trafficking and human disease
Abstract
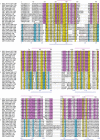
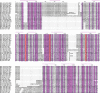
The Sec7 domain ADP-ribosylation factor (Arf) guanine nucleotide exchange factors (GEFs) are found in all eukaryotes, and are involved in membrane remodeling processes throughout the cell. This review is focused on members of the GBF/Gea and BIG/Sec7 subfamilies of Arf GEFs, all of which use the class I Arf proteins (Arf1-3) as substrates, and play a fundamental role in trafficking in the endoplasmic reticulum (ER)-Golgi and endosomal membrane systems. Members of the GBF/Gea and BIG/Sec7 subfamilies are large proteins on the order of 200 kDa, and they possess multiple homology domains. Phylogenetic analyses indicate that both of these subfamilies of Arf GEFs have members in at least five out of the six eukaryotic supergroups, and hence were likely present very early in eukaryotic evolution. The homology domains of the large Arf1 GEFs play important functional roles, and are involved in interactions with numerous protein partners. The large Arf1 GEFs have been implicated in several human diseases. They are crucial host factors for the replication of several viral pathogens, including poliovirus, coxsackievirus, mouse hepatitis coronavirus, and hepatitis C virus. Mutations in the BIG2 Arf1 GEF have been linked to autosomal recessive periventricular heterotopia, a disorder of neuronal migration that leads to severe malformation of the cerebral cortex. Understanding the roles of the Arf1 GEFs in membrane dynamics is crucial to a full understanding of trafficking in the secretory and endosomal pathways, which in turn will provide essential insights into human diseases that arise from misregulation of these pathways.
Figures








Similar articles
-
A single class of ARF GTPase activated by several pathway-specific ARF-GEFs regulates essential membrane traffic in Arabidopsis.PLoS Genet. 2018 Nov 15;14(11):e1007795. doi: 10.1371/journal.pgen.1007795. eCollection 2018 Nov. PLoS Genet. 2018. PMID: 30439956 Free PMC article.
-
ARNO3, a Sec7-domain guanine nucleotide exchange factor for ADP ribosylation factor 1, is involved in the control of Golgi structure and function.Proc Natl Acad Sci U S A. 1998 Aug 18;95(17):9926-31. doi: 10.1073/pnas.95.17.9926. Proc Natl Acad Sci U S A. 1998. PMID: 9707577 Free PMC article.
-
Specificities for the small G proteins ARF1 and ARF6 of the guanine nucleotide exchange factors ARNO and EFA6.J Biol Chem. 2001 Jul 6;276(27):24925-30. doi: 10.1074/jbc.M103284200. Epub 2001 May 7. J Biol Chem. 2001. PMID: 11342560
-
Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors.Trends Cell Biol. 2000 Feb;10(2):60-7. doi: 10.1016/s0962-8924(99)01699-2. Trends Cell Biol. 2000. PMID: 10652516 Review.
-
Arf, Sec7 and Brefeldin A: a model towards the therapeutic inhibition of guanine nucleotide-exchange factors.Biochem Soc Trans. 2005 Dec;33(Pt 6):1265-8. doi: 10.1042/BST0331265. Biochem Soc Trans. 2005. PMID: 16246094 Review.
Cited by
-
The Arf-GAP Age2 localizes to the late-Golgi via a conserved amphipathic helix.bioRxiv [Preprint]. 2023 Jul 24:2023.07.23.550229. doi: 10.1101/2023.07.23.550229. bioRxiv. 2023. Update in: Mol Biol Cell. 2023 Nov 1;34(12):ar119. doi: 10.1091/mbc.E23-07-0283. PMID: 37546741 Free PMC article. Updated. Preprint.
-
Origin and evolution of fungal HECT ubiquitin ligases.Sci Rep. 2018 Apr 23;8(1):6419. doi: 10.1038/s41598-018-24914-x. Sci Rep. 2018. PMID: 29686411 Free PMC article.
-
The Arf GEF GBF1 and Arf4 synergize with the sensory receptor cargo, rhodopsin, to regulate ciliary membrane trafficking.J Cell Sci. 2017 Dec 1;130(23):3975-3987. doi: 10.1242/jcs.205492. Epub 2017 Oct 12. J Cell Sci. 2017. PMID: 29025970 Free PMC article.
-
Structural Insights into Arl1-Mediated Targeting of the Arf-GEF BIG1 to the trans-Golgi.Cell Rep. 2016 Jul 19;16(3):839-50. doi: 10.1016/j.celrep.2016.06.022. Epub 2016 Jun 30. Cell Rep. 2016. PMID: 27373159 Free PMC article.
-
(+)RNA viruses rewire cellular pathways to build replication organelles.Curr Opin Virol. 2012 Dec;2(6):740-7. doi: 10.1016/j.coviro.2012.09.006. Epub 2012 Oct 1. Curr Opin Virol. 2012. PMID: 23036609 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

