De novo molecular modeling and biophysical characterization of Manduca sexta eclosion hormone
- PMID: 19670911
- PMCID: PMC2792118
- DOI: 10.1021/bi901078y
De novo molecular modeling and biophysical characterization of Manduca sexta eclosion hormone
Abstract
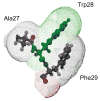
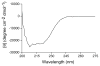
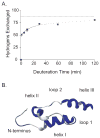
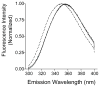
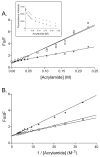
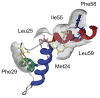
Eclosion hormone (EH) is an integral component in the cascade regulating the behaviors culminating in emergence of an insect from its old exoskeleton. Little is known regarding the EH solution structure; consequently, we utilized a computational approach to generate a hypothetical structure for Manduca sexta EH. The de novo algorithm exploited the restricted conformational space of disulfide bonds (Cys14-Cys38, Cys18-Cys34, and Cys21-Cys49) and predicted secondary structure elements to generate a thermodynamically stable structure characterized by 55% helical content, an unstructured N-terminus, a helical C-terminus, and a solvent-exposed loop containing Trp28 and Phe29. Both the strain and pseudo energies of the predicted peptide compare favorably with those of known structures. The 62-amino acid peptide was synthesized, folded, assayed for activity, and structurally characterized to confirm the validity of the model. The helical content is supported by circular dichroism and hydrogen-deuterium exchange mass spectrometry. Fluorescence emission spectra and acrylamide quenching are consistent with the solvent exposure predicted for Trp28, which is shielded by Phe29. Furthermore, thermodynamically stable conformations that deviated only slightly from the predicted Manduca EH structure were generated in silico for the Bombyx mori and Drosophila melanogaster EHs, indicating that the conformation is not species-dependent. In addition, the biological activities of known mutants and deletion peptides were rationalized with the predicted Manduca EH structure, and we found that, on the basis of sequence conservation, functionally important residues map to two conserved hydrophobic clusters incorporating the C-terminus and the first loop.
Figures




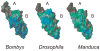

Similar articles
-
Complete structure of eclosion hormone of Manduca sexta. Assignment of disulfide bond location.Int J Pept Protein Res. 1992 Jan;39(1):29-35. doi: 10.1111/j.1399-3011.1992.tb01552.x. Int J Pept Protein Res. 1992. PMID: 1634328
-
The functional residues and their representation by a hypothetical 3D model of silkworm eclosion hormone.Protein Eng. 1998 Sep;11(9):769-73. doi: 10.1093/protein/11.9.769. Protein Eng. 1998. PMID: 9796825
-
Proteolytic activation and function of the cytokine Spätzle in the innate immune response of a lepidopteran insect, Manduca sexta.FEBS J. 2010 Jan;277(1):148-62. doi: 10.1111/j.1742-4658.2009.07465.x. Epub 2009 Nov 26. FEBS J. 2010. PMID: 19968713 Free PMC article.
-
Molecular characterization of a cDNA from the silk moth Bombyx mori encoding Manduca sexta allatotropin peptide.Zoolog Sci. 2002 Mar;19(3):287-92. doi: 10.2108/zsj.19.287. Zoolog Sci. 2002. PMID: 12125926
-
Isolation, characterization and expression of the eclosion hormone gene of Drosophila melanogaster.Eur J Biochem. 1993 Jul 15;215(2):221-8. doi: 10.1111/j.1432-1033.1993.tb18026.x. Eur J Biochem. 1993. PMID: 8344291
Cited by
-
Key Neuropeptides Regulating Molting in Pacific White Shrimp (Penaeus vannamei): Insights from Transcriptomic Analysis.Animals (Basel). 2025 Feb 13;15(4):540. doi: 10.3390/ani15040540. Animals (Basel). 2025. PMID: 40003023 Free PMC article.
-
In silico Neuropeptidome of Female Macrobrachium rosenbergii Based on Transcriptome and Peptide Mining of Eyestalk, Central Nervous System and Ovary.PLoS One. 2015 May 29;10(5):e0123848. doi: 10.1371/journal.pone.0123848. eCollection 2015. PLoS One. 2015. PMID: 26023789 Free PMC article.
References
-
- Truman JW. Hormonal control of insect ecdysis: endocrine cascades for coordinating behavior with physiology. Vitam Horm. 2005;73:1–30. - PubMed
-
- Truman JW, Taghert PH, Copenhaver PF, Tublitz NJ, Schwartz LM. Eclosion hormone may control all ecdysis in insects. Nature. 1981;291:70–71.
-
- Truman JW. The eclosion hormone system of insects. Prog Brain Res. 1992;92:361–374. - PubMed
-
- Baker JD, McNabb SL, Truman JW. The hormonal coordination of behavior and physiology at adult ecdysis in Drosophila melanogaster. J Exp Biol. 1999;202:3037–3048. - PubMed
-
- McNabb SL, Baker JD, Agapite J, Steller H, Riddiford LM, Truman JW. Disruption of a behavioral sequence by targeted death of peptidergic neurons in Drosophila. Neuron. 1997;19:813–823. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

