Meayamycin inhibits pre-messenger RNA splicing and exhibits picomolar activity against multidrug-resistant cells
- PMID: 19671752
- PMCID: PMC2762647
- DOI: 10.1158/1535-7163.MCT-09-0051
Meayamycin inhibits pre-messenger RNA splicing and exhibits picomolar activity against multidrug-resistant cells
Abstract
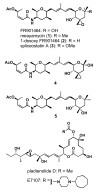
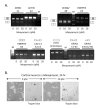
FR901464 is a potent antitumor natural product that binds to the splicing factor 3b complex and inhibits pre-mRNA splicing. Its analogue, meayamycin, is two orders of magnitude more potent as an antiproliferative agent against human breast cancer MCF-7 cells. Here, we report the picomolar antiproliferative activity of meayamycin against various cancer cell lines and multidrug-resistant cells. Time-dependence studies implied that meayamycin may form a covalent bond with its target protein(s). Meayamycin inhibited pre-mRNA splicing in HEK-293 cells but not alternative splicing in a neuronal system. Meayamycin exhibited specificity toward human lung cancer cells compared with nontumorigenic human lung fibroblasts and retained picomolar growth-inhibitory activity against multidrug-resistant cells. These data suggest that meayamycin is a useful chemical probe to study pre-mRNA splicing in live cells and is a promising lead as an anticancer agent.
Conflict of interest statement
Figures




References
-
- Nakajima H, Sato B, Fujita T, Takase S, Terano H, Okuhara M. New antitumor substances, FR901463, FR901464 and FR901465. 1. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J Antibiot. 1996;49:1196–203. - PubMed
-
- Nakajima H, Hori Y, Terano H, et al. New antitumor substances, FR901463, FR901464 and FR901465. II. Activities against experimental tumors in mice and mechanism of action. J Antibiot. 1996;49:1204–11. - PubMed
-
- Thompson CF, Jamison TF, Jacobsen EN. Total synthesis of FR901464. Convergent assembly of chiral components prepared by asymmetric catalysis. J Am Chem Soc. 2000;122:10482–3.
-
- Horigome M, Motoyoshi H, Watanabe H, Kitahara T. A synthesis of FR901464. Tetrahedron Letters. 2001;42:8207–10.
-
- Thompson CF, Jamison TF, Jacobsen EN. FR901464: total synthesis, proof of structure, and evaluation of synthetic analogues. J Am Chem Soc. 2001;123:9974–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

