The Wnt receptor, Lrp5, is expressed by mouse mammary stem cells and is required to maintain the basal lineage
- PMID: 19672307
- PMCID: PMC2720450
- DOI: 10.1371/journal.pone.0006594
The Wnt receptor, Lrp5, is expressed by mouse mammary stem cells and is required to maintain the basal lineage
Abstract
Background: Ectopic Wnt signaling induces increased stem/progenitor cell activity in the mouse mammary gland, followed by tumor development. The Wnt signaling receptors, Lrp5/6, are uniquely required for canonical Wnt activity. Previous data has shown that the absence of Lrp5 confers resistance to Wnt1-induced tumor development.
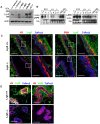
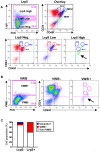
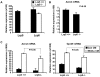
Methodology/principal findings: Here, we show that all basal mammary cells express Lrp5, and co-express Lrp6 in a similar fashion. Though Wnt dependent transcription of key target genes is relatively unchanged in mammary epithelial cell cultures, the absence of Lrp5 specifically depletes adult regenerative stem cell activity (to less than 1%). Stem cell activity can be enriched by >200 fold (over 80% of activity), based on high Lrp5 expression alone. Though Lrp5 null glands have apparent normal function, the basal lineage is relatively reduced (from 42% basal/total epithelial cells to 22%) and Lrp5-/- mammary epithelial cells show enhanced expression of senescence-associated markers in vitro, as measured by expression of p16(Ink4a) and TA-p63.
Conclusions/significance: This is the first single biomarker that has been demonstrated to be functionally involved in stem cell maintenance. Together, these results demonstrate that Wnt signaling through Lrp5 is an important component of normal mammary stem cell function.
Conflict of interest statement
Figures




References
-
- Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–398. - PubMed
-
- Brennan KR, Brown AM. Wnt proteins in mammary development and cancer. J Mammary Gland Biol Neoplasia. 2004;9:119–131. - PubMed
-
- Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. - PubMed
-
- Huelsken J, Birchmeier W. New aspects of Wnt signaling pathways in higher vertebrates. Curr Opin Genet Dev. 2001;11:547–553. - PubMed
-
- Katoh M. WNT signaling in stem cell biology and regenerative medicine. Curr Drug Targets. 2008;9:565–570. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

