Multiple antibiotic resistance in Arabidopsis is conferred by mutations in a chloroplast-localized transport protein
- PMID: 19675150
- PMCID: PMC2754617
- DOI: 10.1104/pp.109.143487
Multiple antibiotic resistance in Arabidopsis is conferred by mutations in a chloroplast-localized transport protein
Abstract
Widespread antibiotic resistance is a major public health concern, and plants represent an emerging antibiotic exposure route. Recent studies indicate that crop plants fertilized with antibiotic-laden animal manure accumulate antibiotics; however, the molecular mechanisms of antibiotic entry and subcellular partitioning within plant cells remain unknown. Here, we report that mutations in the Arabidopsis (Arabidopsis thaliana) locus Multiple Antibiotic Resistance1 (MAR1) confer resistance, while MAR1 overexpression causes hypersensitivity to multiple aminoglycoside antibiotics. Additionally, yeast expressing MAR1 are hypersensitive to the aminoglycoside G418. MAR1 encodes a protein with 11 putative transmembrane domains with low similarity to ferroportin1 from Danio rerio. A MAR1:yellow fluorescent protein fusion localizes to the chloroplast, and chloroplasts from plants overexpressing MAR1 accumulate more of the aminoglycoside gentamicin, while mar1-1 mutant chloroplasts accumulate less than the wild type. MAR1 overexpression lines are slightly chlorotic, and chlorosis is rescued by exogenous iron. MAR1 expression is also down-regulated by low iron. These data suggest that MAR1 is a plastid transporter that is likely to be involved in cellular iron homeostasis and allows opportunistic entry of multiple antibiotics into the chloroplast.
Figures
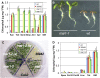

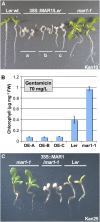
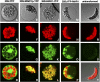
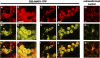
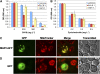
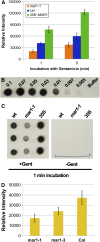
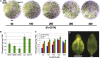

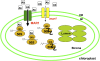
Comment in
-
The MAR1 transporter is an opportunistic entry point for antibiotics.Plant Signal Behav. 2010 Jan;5(1):49-52. doi: 10.4161/psb.5.1.10142. Plant Signal Behav. 2010. PMID: 20592808 Free PMC article.
References
-
- Aronsson H, Jarvis P (2002) A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett 529: 215–220 - PubMed
-
- Aufsatz W, Nehlin L, Voronin V, Schmidt A, Matzke A, Matzke M (2009) A novel strategy for obtaining kanamycin resistance in Arabidopsis thaliana by silencing an endogenous gene encoding a putative chloroplast transporter. Biotechnol J 4: 224–229 - PubMed
-
- Boxall A, Johnson P, Smith E, Sinclair C, Stutt E, Levy L (2006) Uptake of veterinary medicines from soils into plants. J Agric Food Chem 54: 2288–2297 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

