Unraveling the evolution of cytokinin signaling
- PMID: 19675156
- PMCID: PMC2754637
- DOI: 10.1104/pp.109.139188
Unraveling the evolution of cytokinin signaling
Abstract
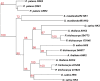
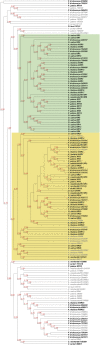
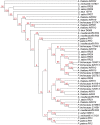
The conquest of the land by plants required dramatic morphological and metabolic adaptations. Complex developmental programs under tight regulation evolved during this process. Key regulators of plant development are phytohormones, such as cytokinins. Cytokinins are adenine derivatives that affect various processes in plants. The cytokinin signal transduction system, which is mediated via a multistep variant of the bacterial two-component signaling system, is well characterized in the model plant Arabidopsis (Arabidopsis thaliana). To understand the origin and evolutionary pattern of this signaling pathway, we surveyed the genomes of several sequenced key plant species ranging from unicellular algae, moss, and lycophytes, to higher land plants, including Arabidopsis and rice (Oryza sativa), for proteins involved in cytokinin signal transduction. Phylogenetic analysis revealed that the hormone-binding receptor and a class of negative regulators first appeared in land plants. Other components of the signaling pathway were present in all species investigated. Furthermore, we found that the receptors evolved under different evolutionary constraints from the other components of the pathway: The number of receptors remained fairly constant, while the other protein families expanded.
Figures




Similar articles
-
CHASE domain-containing receptors play an essential role in the cytokinin response of the moss Physcomitrella patens.J Exp Bot. 2016 Feb;67(3):667-79. doi: 10.1093/jxb/erv479. Epub 2015 Nov 23. J Exp Bot. 2016. PMID: 26596764 Free PMC article.
-
A subfamily of putative cytokinin receptors is revealed by an analysis of the evolution of the two-component signaling system of plants.Plant Physiol. 2014 May;165(1):227-37. doi: 10.1104/pp.113.228080. Epub 2014 Feb 11. Plant Physiol. 2014. PMID: 24520157 Free PMC article.
-
The more, the merrier: cytokinin signaling beyond Arabidopsis.Plant Signal Behav. 2010 Nov;5(11):1384-90. doi: 10.4161/psb.5.11.13157. Epub 2010 Nov 1. Plant Signal Behav. 2010. PMID: 21045560 Free PMC article. Review.
-
Members of a recently discovered subfamily of cytokinin receptors display differences and similarities to their classical counterparts.Plant Signal Behav. 2015;10(2):e984512. doi: 10.4161/21659087.2014.984512. Plant Signal Behav. 2015. PMID: 25826259 Free PMC article.
-
Updates on the model and the evolution of cytokinin signaling.Curr Opin Plant Biol. 2013 Oct;16(5):569-74. doi: 10.1016/j.pbi.2013.09.001. Epub 2013 Sep 28. Curr Opin Plant Biol. 2013. PMID: 24080474 Review.
Cited by
-
The structure of B-ARR reveals the molecular basis of transcriptional activation by cytokinin.Proc Natl Acad Sci U S A. 2024 Jan 16;121(3):e2319335121. doi: 10.1073/pnas.2319335121. Epub 2024 Jan 10. Proc Natl Acad Sci U S A. 2024. PMID: 38198526 Free PMC article.
-
New Insight into HPts as Hubs in Poplar Cytokinin and Osmosensing Multistep Phosphorelays: Cytokinin Pathway Uses Specific HPts.Plants (Basel). 2019 Dec 11;8(12):591. doi: 10.3390/plants8120591. Plants (Basel). 2019. PMID: 31835814 Free PMC article.
-
Structure and character analysis of cotton response regulator genes family reveals that GhRR7 responses to draught stress.Biol Res. 2022 Aug 16;55(1):27. doi: 10.1186/s40659-022-00394-2. Biol Res. 2022. PMID: 35974357 Free PMC article.
-
Advances in upstream players of cytokinin phosphorelay: receptors and histidine phosphotransfer proteins.Plant Cell Rep. 2012 May;31(5):789-99. doi: 10.1007/s00299-012-1229-9. Epub 2012 Feb 15. Plant Cell Rep. 2012. PMID: 22350315 Review.
-
Functional roles of three cutin biosynthetic acyltransferases in cytokinin responses and skotomorphogenesis.PLoS One. 2015 Mar 24;10(3):e0121943. doi: 10.1371/journal.pone.0121943. eCollection 2015. PLoS One. 2015. PMID: 25803274 Free PMC article.
References
-
- Anantharaman V, Aravind L (2001) The CHASE domain: a predicted ligand-binding module in plant cytokinin receptors and other eukaryotic and bacterial receptors. Trends Biochem Sci 26: 579–582 - PubMed
-
- Anantharaman V, Iyer LM, Aravind L (2007) Comparative genomics of protists: new insights into the evolution of eukaryotic signal transduction and gene regulation. Annu Rev Microbiol 61: 453–475 - PubMed
-
- Anjard C, Loomis WF (2008) Cytokinins induce sporulation in Dictyostelium. Development 135: 819–827 - PubMed
-
- Archibald JM (2006) Algal genomics: exploring the imprint of endosymbiosis. Curr Biol 16: 1033–1035 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

