Unraveling the evolution of cytokinin signaling
- PMID: 19675156
- PMCID: PMC2754637
- DOI: 10.1104/pp.109.139188
Unraveling the evolution of cytokinin signaling
Abstract
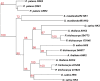
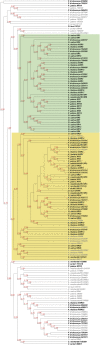
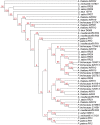
The conquest of the land by plants required dramatic morphological and metabolic adaptations. Complex developmental programs under tight regulation evolved during this process. Key regulators of plant development are phytohormones, such as cytokinins. Cytokinins are adenine derivatives that affect various processes in plants. The cytokinin signal transduction system, which is mediated via a multistep variant of the bacterial two-component signaling system, is well characterized in the model plant Arabidopsis (Arabidopsis thaliana). To understand the origin and evolutionary pattern of this signaling pathway, we surveyed the genomes of several sequenced key plant species ranging from unicellular algae, moss, and lycophytes, to higher land plants, including Arabidopsis and rice (Oryza sativa), for proteins involved in cytokinin signal transduction. Phylogenetic analysis revealed that the hormone-binding receptor and a class of negative regulators first appeared in land plants. Other components of the signaling pathway were present in all species investigated. Furthermore, we found that the receptors evolved under different evolutionary constraints from the other components of the pathway: The number of receptors remained fairly constant, while the other protein families expanded.
Figures




References
-
- Anantharaman V, Aravind L (2001) The CHASE domain: a predicted ligand-binding module in plant cytokinin receptors and other eukaryotic and bacterial receptors. Trends Biochem Sci 26: 579–582 - PubMed
-
- Anantharaman V, Iyer LM, Aravind L (2007) Comparative genomics of protists: new insights into the evolution of eukaryotic signal transduction and gene regulation. Annu Rev Microbiol 61: 453–475 - PubMed
-
- Anjard C, Loomis WF (2008) Cytokinins induce sporulation in Dictyostelium. Development 135: 819–827 - PubMed
-
- Archibald JM (2006) Algal genomics: exploring the imprint of endosymbiosis. Curr Biol 16: 1033–1035 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

