Analysis of PHOTOPERIOD SENSITIVITY5 sheds light on the role of phytochromes in photoperiodic flowering in rice
- PMID: 19675157
- PMCID: PMC2754645
- DOI: 10.1104/pp.109.139097
Analysis of PHOTOPERIOD SENSITIVITY5 sheds light on the role of phytochromes in photoperiodic flowering in rice
Abstract
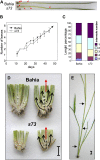
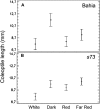
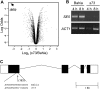
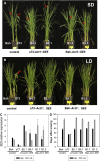
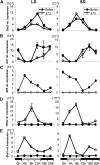
A great number of plants synchronize flowering with day length. In rice (Oryza sativa), photoperiod is the primary environmental cue that triggers flowering. Here, we show that the s73 mutant, identified in a gamma-irradiated Bahia collection, displays early flowering and photoperiodic insensitivity due to a null mutation in the PHOTOPERIOD SENSITIVITY5 (SE5) gene, which encodes an enzyme implicated in phytochrome chromophore biosynthesis. s73 mutant plants show a number of alterations in the characteristic diurnal expression patterns of master genes involved in photoperiodic control of flowering, resulting in up-regulation of the floral integrator Heading date3a (Hd3a). Early heading date1 (Ehd1), an additional rice floral activator, was also highly expressed in the s73 mutant, suggesting that SE5 represses Ehd1 in wild-type plants. Silencing of Ehd1 in both Bahia and s73 backgrounds indicated that SE5 regulates Ehd1 expression. The data also indicate that SE5 confers photoperiodic sensitivity through regulation of Hd1. These results provide direct evidence that phytochromes inhibit flowering by affecting both Hd1 and Ehd1 flowering pathways.
Figures


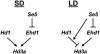
References
-
- Biswas KK, Neumann R, Haga K, Yatoh O, Iino M (2003) Photomorphogenesis of rice seedlings: a mutant impaired in phytochrome-mediated inhibition of coleoptile growth. Plant Cell Physiol 44: 242–254 - PubMed
-
- Conesa A, Gotees S, García-Gómez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal annotation and visualization tool in functional genomics research. Application note. Bioinformatics 21: 3674–3676 - PubMed
-
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 - PubMed
-
- Domingo C, Andrés F, Iglesias D, Tharreau D, Talón M (2009) Depletion of auxin content mediated by OsGH3.1 overexpression activates defense response and enhances resistance to fungal pathogen in rice. Mol Plant Microbe Interact 22: 201–210 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

