Design of functional metalloproteins
- PMID: 19675646
- PMCID: PMC2770889
- DOI: 10.1038/nature08304
Design of functional metalloproteins
Abstract
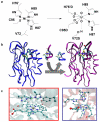
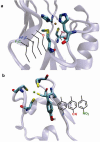
Metalloproteins catalyse some of the most complex and important processes in nature, such as photosynthesis and water oxidation. An ultimate test of our knowledge of how metalloproteins work is to design new metalloproteins. Doing so not only can reveal hidden structural features that may be missing from studies of native metalloproteins and their variants, but also can result in new metalloenzymes for biotechnological and pharmaceutical applications. Although it is much more challenging to design metalloproteins than non-metalloproteins, much progress has been made in this area, particularly in functional design, owing to recent advances in areas such as computational and structural biology.
Figures


References
-
- Lu Y, Berry SM, Pfister TD. Engineering novel metalloproteins: design of metal-binding sites into native protein scaffolds. Chem. Rev. 2001;101:3047–3080. - PubMed
-
- Barker PD. Designing redox metalloproteins from bottom-up and top-down perspectives. Curr. Opin. Struct. Biol. 2003;13:490–499. - PubMed
-
- DeGrado WF, Summa CM, Pavone V, Nastri F, Lombardi A. De novo design and structural characterization of proteins and metalloproteins. Annu. Rev. Biochem. 1999;68:779–819. - PubMed
-
- Reedy CJ, Gibney BR. Heme protein assemblies. Chem. Rev. 2004;104:617–649. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

