The origin of allosteric functional modulation: multiple pre-existing pathways
- PMID: 19679084
- PMCID: PMC2749652
- DOI: 10.1016/j.str.2009.06.008
The origin of allosteric functional modulation: multiple pre-existing pathways
Abstract
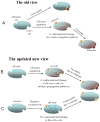
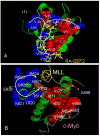
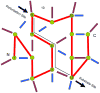
Although allostery draws increasing attention, not much is known about allosteric mechanisms. Here we argue that in all proteins, allosteric signals transmit through multiple, pre-existing pathways; which pathways dominate depend on protein topologies, specific binding events, covalent modifications, and cellular (environmental) conditions. Further, perturbation events at any site on the protein surface (or in the interior) will not create new pathways but only shift the pre-existing ensemble of pathways. Drugs binding at different sites or mutational events in disease shift the ensemble toward the same conformations; however, the relative populations of the different states will change. Consequently the observed functional, conformational, and dynamic effects will be different. This is the origin of allosteric functional modulation in dynamic proteins: allostery does not necessarily need to invoke conformational rearrangements to control protein activity and pre-existing pathways are always defaulted to during allostery regardless of the stimulant and perturbation site in the protein.
Figures
References
-
- Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, et al. The Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2002;58:899–907. - PubMed
-
- Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–1642. - PubMed
-
- Bogoyevitch MA, Fairlie DP. A new paradigm for protein kinase inhibition: blocking phosphorylation without directly targeting ATP binding. Drug Discov Today. 2007;12:622–633. - PubMed
-
- Bruschweiler S, Schanda P, Kloiber K, Brutscher B, Kontaxis G, Konrat R, Tollinger M. Direct Observation of the Dynamic Process Underlying Allosteric Signal Transmission. J Am Chem Soc. 2009;131:3063–3068. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

