The CATH hierarchy revisited-structural divergence in domain superfamilies and the continuity of fold space
- PMID: 19679085
- PMCID: PMC2741583
- DOI: 10.1016/j.str.2009.06.015
The CATH hierarchy revisited-structural divergence in domain superfamilies and the continuity of fold space
Abstract
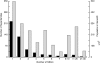
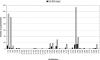
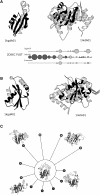
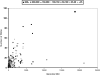
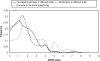
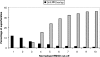
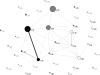
This paper explores the structural continuum in CATH and the extent to which superfamilies adopt distinct folds. Although most superfamilies are structurally conserved, in some of the most highly populated superfamilies (4% of all superfamilies) there is considerable structural divergence. While relatives share a similar fold in the evolutionary conserved core, diverse elaborations to this core can result in significant differences in the global structures. Applying similar protocols to examine the extent to which structural overlaps occur between different fold groups, it appears this effect is confined to just a few architectures and is largely due to small, recurring super-secondary motifs (e.g., alphabeta-motifs, alpha-hairpins). Although 24% of superfamilies overlap with superfamilies having different folds, only 14% of nonredundant structures in CATH are involved in overlaps. Nevertheless, the existence of these overlaps suggests that, in some regions of structure space, the fold universe should be seen as more continuous.
Figures


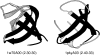
References
-
- Chandonia J.M., Brenner S.E. Implications of structural genomics target selection strategies: Pfam5000, whole genome, and random approaches. Proteins. 2005;58:166–179. - PubMed
-
- Dengler U., Siddiqui A.S., Barton G.J. Protein structural domains: analysis of the 3Dee domains database. Proteins. 2001;42:332–344. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

