Pervasive transcription constitutes a new level of eukaryotic genome regulation
- PMID: 19680288
- PMCID: PMC2750061
- DOI: 10.1038/embor.2009.181
Pervasive transcription constitutes a new level of eukaryotic genome regulation
Abstract
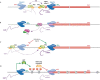
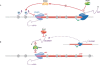
During the past few years, it has become increasingly evident that the expression of eukaryotic genomes is far more complex than had been previously noted. The idea that the transcriptome is derived exclusively from protein-coding genes and some specific non-coding RNAs--such as snRNAs, snoRNAs, tRNAs or rRNAs--has been swept away by numerous studies indicating that RNA polymerase II can be found at almost any genomic location. Pervasive transcription is widespread and, far from being a futile process, has a crucial role in controlling gene expression and genomic plasticity. Here, we review recent findings that point to cryptic transcription as a fundamental component of the regulation of eukaryotic genomes.
Figures
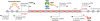

References
-
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ (2007) Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747 - PubMed
-
- Arigo JT, Eyler DE, Carroll KL, Corden JL (2006) Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell 23: 841–851 - PubMed
-
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

