A condensation-ordering mechanism in nanoparticle-catalyzed peptide aggregation
- PMID: 19680431
- PMCID: PMC2715216
- DOI: 10.1371/journal.pcbi.1000458
A condensation-ordering mechanism in nanoparticle-catalyzed peptide aggregation
Abstract
Nanoparticles introduced in living cells are capable of strongly promoting the aggregation of peptides and proteins. We use here molecular dynamics simulations to characterise in detail the process by which nanoparticle surfaces catalyse the self-assembly of peptides into fibrillar structures. The simulation of a system of hundreds of peptides over the millisecond timescale enables us to show that the mechanism of aggregation involves a first phase in which small structurally disordered oligomers assemble onto the nanoparticle and a second phase in which they evolve into highly ordered as their size increases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

 microseconds, the peptides are in their monomeric state. (B) At intermediate times,
microseconds, the peptides are in their monomeric state. (B) At intermediate times,  milliseconds, small oligomeric assemblies form on the nanoparticle surface. (C) At later times,
milliseconds, small oligomeric assemblies form on the nanoparticle surface. (C) At later times,  milliseconds, these oligomers re-order into fibrillar structures as their size increases. Peptides that do not form intermolecular hydrogen bonds are shown in blue, while peptides that form intermolecular hydrogen bonds are assigned a random colour, which is the same for peptides that belong to the same
milliseconds, these oligomers re-order into fibrillar structures as their size increases. Peptides that do not form intermolecular hydrogen bonds are shown in blue, while peptides that form intermolecular hydrogen bonds are assigned a random colour, which is the same for peptides that belong to the same  . Two peptides are defined as belonging to the same cluster if their centres of mass distance is less than 5 Å. Two peptides are taken to participate within a
. Two peptides are defined as belonging to the same cluster if their centres of mass distance is less than 5 Å. Two peptides are taken to participate within a  if they form more than four inter-chain hydrogen bonds with each other. The spherical nanoparticle is displayed in orange in the centre of the simulation box; the diameter of the peptides is slightly reduced for illustration purposes. Panels (B) and (C) show enlarged views of the nanoparticle-peptide system. The simulation was performed at
if they form more than four inter-chain hydrogen bonds with each other. The spherical nanoparticle is displayed in orange in the centre of the simulation box; the diameter of the peptides is slightly reduced for illustration purposes. Panels (B) and (C) show enlarged views of the nanoparticle-peptide system. The simulation was performed at  ,
,  ,
,  , and
, and  .
.
 observed during a simulation in presence of a hard sphere nanoparticle:
observed during a simulation in presence of a hard sphere nanoparticle:  ,
,  (green line), and several hydrophobic nanoparticles that differ in diameter and hydrophobicity:
(green line), and several hydrophobic nanoparticles that differ in diameter and hydrophobicity:  ,
,  (blue line),
(blue line),  ,
,  (red line), and
(red line), and  ,
,  (black line). The results are averaged over ten independent simulation runs and the error bars correspond to the standard deviation of the mean. (B) Number of clusters
(black line). The results are averaged over ten independent simulation runs and the error bars correspond to the standard deviation of the mean. (B) Number of clusters  of size
of size  as a function of time:
as a function of time:  microseconds (left),
microseconds (left),  milliseconds (middle),
milliseconds (middle),  milliseconds (right), for the MD trajectory and parameters described in Fig. 1. Black lines correspond to all clusters formed in the system; red lines correspond to the number of clusters formed on the nanoparticle surface. (C) Structural order parameter
milliseconds (right), for the MD trajectory and parameters described in Fig. 1. Black lines correspond to all clusters formed in the system; red lines correspond to the number of clusters formed on the nanoparticle surface. (C) Structural order parameter  as a function of the cluster size
as a function of the cluster size  averaged over ten independent simulations. The line colours are as described in (A). (D) Normalized density profile
averaged over ten independent simulations. The line colours are as described in (A). (D) Normalized density profile  , where
, where  is the bulk density of the system, as a function of the distance from the centre of mass of the nanoparticle at the beginning of the simulation,
is the bulk density of the system, as a function of the distance from the centre of mass of the nanoparticle at the beginning of the simulation,  microseconds (left panel), intermediate times,
microseconds (left panel), intermediate times,  milliseconds (middle panel), and at the end
milliseconds (middle panel), and at the end  milliseconds (right panel). The different line colours are as described in (A) and correspond to the different seed sizes and peptide seed interaction energies. The results are averaged over ten independent simulations and the error bars correspond to the standard deviation of the mean.
milliseconds (right panel). The different line colours are as described in (A) and correspond to the different seed sizes and peptide seed interaction energies. The results are averaged over ten independent simulations and the error bars correspond to the standard deviation of the mean.
 ,
,  at
at  microseconds(left),
microseconds(left),  milliseconds (middle),
milliseconds (middle),  milliseconds (right). (B)
milliseconds (right). (B)  ,
,  at
at  microseconds (left),
microseconds (left),  milliseconds (middle),
milliseconds (middle),  milliseconds (right). The concentration and temperature are
milliseconds (right). The concentration and temperature are  ,
,  respectively, and the colour code is as described in Fig 1.
respectively, and the colour code is as described in Fig 1.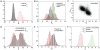
 and
and  between
between  atoms
atoms  and
and  used to define a
used to define a  helical hydrogen bond assigned to atoms
helical hydrogen bond assigned to atoms  with
with  . (B) Histogram of distances
. (B) Histogram of distances  between
between  atoms
atoms  that form a parallel, anti-parallel, or helical hydrogen bond. For the
that form a parallel, anti-parallel, or helical hydrogen bond. For the  helical hydrogen bond
helical hydrogen bond  . (C) Illustration of the alternation of distances for consecutive
. (C) Illustration of the alternation of distances for consecutive  atoms that form anti-parallel hydrogen bonds. (D) as in (C). (E) The distance
atoms that form anti-parallel hydrogen bonds. (D) as in (C). (E) The distance  is used to define hydrogen bonds between atoms
is used to define hydrogen bonds between atoms  in parallel and anti parallel
in parallel and anti parallel  . (F) Distances used to define cooperative hydrogen bonds between two consecutive atoms
. (F) Distances used to define cooperative hydrogen bonds between two consecutive atoms  and
and  that form parallel
that form parallel  or
or  and
and  that form anti parallel
that form anti parallel  .
.References
-
- Fischer HC, Chan WCW. Nanotoxicity: the growing need for in vivo study. Curr Opin Biotech. 2007;18:565–571. - PubMed
-
- Lynch I, Dawson KA. Protein-nanoparticle interactions. Nano Today. 2008;3:40–47.
-
- Schulze C, Kroll A, Lehr CM, Schafer UF, Becker K, et al. Not ready to use – overcoming pitfalls when dispersing nanoparticles in physiological media. Nanotoxicology. 2008;2:51–61.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

