Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context
- PMID: 19680444
- PMCID: PMC2718614
- DOI: 10.1371/journal.pgen.1000602
Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context
Abstract
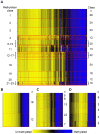
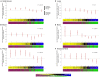
Epigenetic control of gene transcription is critical for normal human development and cellular differentiation. While alterations of epigenetic marks such as DNA methylation have been linked to cancers and many other human diseases, interindividual epigenetic variations in normal tissues due to aging, environmental factors, or innate susceptibility are poorly characterized. The plasticity, tissue-specific nature, and variability of gene expression are related to epigenomic states that vary across individuals. Thus, population-based investigations are needed to further our understanding of the fundamental dynamics of normal individual epigenomes. We analyzed 217 non-pathologic human tissues from 10 anatomic sites at 1,413 autosomal CpG loci associated with 773 genes to investigate tissue-specific differences in DNA methylation and to discern how aging and exposures contribute to normal variation in methylation. Methylation profile classes derived from unsupervised modeling were significantly associated with age (P<0.0001) and were significant predictors of tissue origin (P<0.0001). In solid tissues (n = 119) we found striking, highly significant CpG island-dependent correlations between age and methylation; loci in CpG islands gained methylation with age, loci not in CpG islands lost methylation with age (P<0.001), and this pattern was consistent across tissues and in an analysis of blood-derived DNA. Our data clearly demonstrate age- and exposure-related differences in tissue-specific methylation and significant age-associated methylation patterns which are CpG island context-dependent. This work provides novel insight into the role of aging and the environment in susceptibility to diseases such as cancer and critically informs the field of epigenomics by providing evidence of epigenetic dysregulation by age-related methylation alterations. Collectively we reveal key issues to consider both in the construction of reference and disease-related epigenomes and in the interpretation of potentially pathologically important alterations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Russo V, Martienssen RA, Riggs AD. Cold Spring Harbor Laboratory Press; 1996. Epigenetic mechanisms of gene regulation.
-
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. - PubMed
-
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. - PubMed
-
- Xu GL, Bestor TH, Bourc'his D, Hsieh CL, Tommerup N, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 CA057494/CA/NCI NIH HHS/United States
- R01 ES006717/ES/NIEHS NIH HHS/United States
- R01 CA57494/CA/NCI NIH HHS/United States
- P42 ES007373/ES/NIEHS NIH HHS/United States
- R01 CA100679/CA/NCI NIH HHS/United States
- R01 CA126939/CA/NCI NIH HHS/United States
- R01 CA052689/CA/NCI NIH HHS/United States
- R01 CA126831/CA/NCI NIH HHS/United States
- R01 CA078609/CA/NCI NIH HHS/United States
- R01 CA121147/CA/NCI NIH HHS/United States
- R01 CA52689/CA/NCI NIH HHS/United States
- P50 CA097257/CA/NCI NIH HHS/United States
- P50 097257/PHS HHS/United States
- P20 RR018728/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

