Diversity and strain specificity of plant cell wall degrading enzymes revealed by the draft genome of Ruminococcus flavefaciens FD-1
- PMID: 19680555
- PMCID: PMC2721979
- DOI: 10.1371/journal.pone.0006650
Diversity and strain specificity of plant cell wall degrading enzymes revealed by the draft genome of Ruminococcus flavefaciens FD-1
Abstract
Background: Ruminococcus flavefaciens is a predominant cellulolytic rumen bacterium, which forms a multi-enzyme cellulosome complex that could play an integral role in the ability of this bacterium to degrade plant cell wall polysaccharides. Identifying the major enzyme types involved in plant cell wall degradation is essential for gaining a better understanding of the cellulolytic capabilities of this organism as well as highlighting potential enzymes for application in improvement of livestock nutrition and for conversion of cellulosic biomass to liquid fuels.
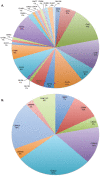
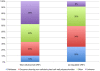
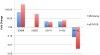
Methodology/principal findings: The R. flavefaciens FD-1 genome was sequenced to 29x-coverage, based on pulsed-field gel electrophoresis estimates (4.4 Mb), and assembled into 119 contigs providing 4,576,399 bp of unique sequence. As much as 87.1% of the genome encodes ORFs, tRNA, rRNAs, or repeats. The GC content was calculated at 45%. A total of 4,339 ORFs was detected with an average gene length of 918 bp. The cellulosome model for R. flavefaciens was further refined by sequence analysis, with at least 225 dockerin-containing ORFs, including previously characterized cohesin-containing scaffoldin molecules. These dockerin-containing ORFs encode a variety of catalytic modules including glycoside hydrolases (GHs), polysaccharide lyases, and carbohydrate esterases. Additionally, 56 ORFs encode proteins that contain carbohydrate-binding modules (CBMs). Functional microarray analysis of the genome revealed that 56 of the cellulosome-associated ORFs were up-regulated, 14 were down-regulated, 135 were unaffected, when R. flavefaciens FD-1 was grown on cellulose versus cellobiose. Three multi-modular xylanases (ORF01222, ORF03896, and ORF01315) exhibited the highest levels of up-regulation.
Conclusions/significance: The genomic evidence indicates that R. flavefaciens FD-1 has the largest known number of fiber-degrading enzymes likely to be arranged in a cellulosome architecture. Functional analysis of the genome has revealed that the growth substrate drives expression of enzymes predicted to be involved in carbohydrate metabolism as well as expression and assembly of key cellulosomal enzyme components.
Conflict of interest statement
Figures



References
-
- Wolin MJ, Miller LT. Interactions of microbial populations in cellulose fermentation. Federation Proceedings. 1983;42:109–113. - PubMed
-
- Sijpesteijn AK. On Ruminococcus flavefaciens, a cellulose decomposing bacterium from the rumen of sheep and cattle. J Gen Microbiol. 1951;5:869–879. - PubMed
-
- Bryant MP. Ruminococcus. In: Sneath PH, Mair HS, Sharpe ME, Holt JG, editors. Bergey's Manual of Systematic Bacteriology. Baltimore: Williams and Wilkins Co; 1986. pp. 1093–1097.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

