A mechanism of transposon-mediated directed mutation
- PMID: 19682247
- PMCID: PMC2973706
- DOI: 10.1111/j.1365-2958.2009.06831.x
A mechanism of transposon-mediated directed mutation
Abstract
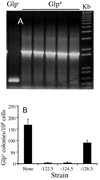
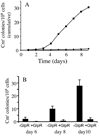
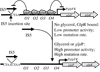
Directed mutation is a proposed process that allows mutations to occur at higher frequencies when they are beneficial. Until now, the existence of such a process has been controversial. Here we describe a novel mechanism of directed mutation mediated by the transposon, IS5 in Escherichia coli. crp deletion mutants mutate specifically to glycerol utilization (Glp(+)) at rates that are enhanced by glycerol or the loss of the glycerol repressor (GlpR), depressed by glucose or glpR overexpression, and RecA-independent. Of the four tandem GlpR binding sites (O1-O4) upstream of the glpFK operon, O4 specifically controls glpFK expression while O1 primarily controls mutation rate in a process mediated by IS5 hopping to a specific site on the E. coli chromosome upstream of the glpFK promoter. IS5 insertion into other gene activation sites is unaffected by the presence of glycerol or the loss of GlpR. The results establish an example of transposon-mediated directed mutation, identify the protein responsible and define the mechanism involved.
Figures





Similar articles
-
Transposon-mediated directed mutation in bacteria and eukaryotes.Front Biosci (Landmark Ed). 2017 Mar 1;22(9):1458-1468. doi: 10.2741/4553. Front Biosci (Landmark Ed). 2017. PMID: 28199212 Free PMC article. Review.
-
Transposon-mediated adaptive and directed mutations and their potential evolutionary benefits.J Mol Microbiol Biotechnol. 2011;21(1-2):59-70. doi: 10.1159/000333108. Epub 2012 Jan 13. J Mol Microbiol Biotechnol. 2011. PMID: 22248543 Free PMC article. Review.
-
Control of Transposon-Mediated Directed Mutation by the Escherichia coli Phosphoenolpyruvate:Sugar Phosphotransferase System.J Mol Microbiol Biotechnol. 2015;25(2-3):226-33. doi: 10.1159/000375375. Epub 2015 Jul 9. J Mol Microbiol Biotechnol. 2015. PMID: 26159081 Free PMC article. Review.
-
Transposon-mediated activation of the Escherichia coli glpFK operon is inhibited by specific DNA-binding proteins: Implications for stress-induced transposition events.Mutat Res. 2016 Nov-Dec;793-794:22-31. doi: 10.1016/j.mrfmmm.2016.10.003. Epub 2016 Oct 27. Mutat Res. 2016. PMID: 27810619 Free PMC article.
-
A novel mechanism of transposon-mediated gene activation.PLoS Genet. 2009 Oct;5(10):e1000689. doi: 10.1371/journal.pgen.1000689. Epub 2009 Oct 16. PLoS Genet. 2009. PMID: 19834539 Free PMC article.
Cited by
-
Zinc-Induced Transposition of Insertion Sequence Elements Contributes to Increased Adaptability of Cupriavidus metallidurans.Front Microbiol. 2016 Mar 23;7:359. doi: 10.3389/fmicb.2016.00359. eCollection 2016. Front Microbiol. 2016. PMID: 27047473 Free PMC article.
-
Disentangling genetic and epigenetic determinants of ultrafast adaptation.Mol Syst Biol. 2016 Dec 15;12(12):892. doi: 10.15252/msb.20166951. Mol Syst Biol. 2016. PMID: 27979908 Free PMC article.
-
The Bacterial Phosphotransferase System: New Frontiers 50 Years after Its Discovery.J Mol Microbiol Biotechnol. 2015;25(2-3):73-8. doi: 10.1159/000381215. Epub 2015 Jul 9. J Mol Microbiol Biotechnol. 2015. PMID: 26159069 Free PMC article.
-
Transposon-mediated directed mutation in bacteria and eukaryotes.Front Biosci (Landmark Ed). 2017 Mar 1;22(9):1458-1468. doi: 10.2741/4553. Front Biosci (Landmark Ed). 2017. PMID: 28199212 Free PMC article. Review.
-
Transposon-mediated directed mutation controlled by DNA binding proteins in Escherichia coli.Front Microbiol. 2014 Aug 1;5:390. doi: 10.3389/fmicb.2014.00390. eCollection 2014. Front Microbiol. 2014. PMID: 25136335 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

