Improved base calling for the Illumina Genome Analyzer using machine learning strategies
- PMID: 19682367
- PMCID: PMC2745764
- DOI: 10.1186/gb-2009-10-8-r83
Improved base calling for the Illumina Genome Analyzer using machine learning strategies
Abstract
The Illumina Genome Analyzer generates millions of short sequencing reads. We present Ibis (Improved base identification system), an accurate, fast and easy-to-use base caller that significantly reduces the error rate and increases the output of usable reads. Ibis is faster and more robust with respect to chemistry and technology than other publicly available packages. Ibis is freely available under the GPL from http://bioinf.eva.mpg.de/Ibis/.
Figures
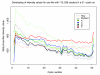
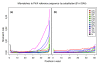

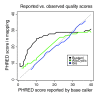
References
-
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. - DOI - PMC - PubMed
-
- R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

