Evolution and functional divergence of NLRP genes in mammalian reproductive systems
- PMID: 19682372
- PMCID: PMC2735741
- DOI: 10.1186/1471-2148-9-202
Evolution and functional divergence of NLRP genes in mammalian reproductive systems
Abstract
Background: NLRPs (Nucleotide-binding oligomerization domain, Leucine rich Repeat and Pyrin domain containing Proteins) are members of NLR (Nod-like receptors) protein family. Recent researches have shown that NLRP genes play important roles in both mammalian innate immune system and reproductive system. Several of NLRP genes were shown to be specifically expressed in the oocyte in mammals. The aim of the present work was to study how these genes evolved and diverged after their duplication, as well as whether natural selection played a role during their evolution.
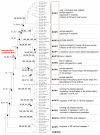
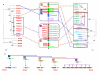
Results: By using in silico methods, we have evaluated the evolution and functional divergence of NLRP genes, in particular of mouse reproduction-related Nlrp genes. We found that (1) major NLRP genes have been duplicated before the divergence of mammals, with certain lineage-specific duplications in primates (NLRP7 and 11) and in rodents (Nlrp1, 4 and 9 duplicates); (2) tandem duplication events gave rise to a mammalian reproduction-related NLRP cluster including NLRP2, 4, 5, 7, 8, 9, 11, 13 and 14 genes; (3) the function of mammalian oocyte-specific NLRP genes (NLRP4, 5, 9 and 14) might have diverged during gene evolution; (4) recent segmental duplications concerning Nlrp4 copies and vomeronasal 1 receptor encoding genes (V1r) have been undertaken in the mouse; and (5) duplicates of Nlrp4 and 9 in the mouse might have been subjected to adaptive evolution.
Conclusion: In conclusion, this study brings us novel information on the evolution of mammalian reproduction-related NLRPs. On the one hand, NLRP genes duplicated and functionally diversified in mammalian reproductive systems (such as NLRP4, 5, 9 and 14). On the other hand, during evolution, different lineages adapted to develop their own NLRP genes, particularly in reproductive function (such as the specific expansion of Nlrp4 and Nlrp9 in the mouse).
Figures
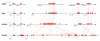
References
-
- Barton GM, Medzhitov R. Toll-like receptors and their ligands. Current topics in microbiology and immunology. 2002;270:81–92. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

