DNA polymerases at the eukaryotic fork-20 years later
- PMID: 19682465
- PMCID: PMC2822129
- DOI: 10.1016/j.mrfmmm.2009.08.002
DNA polymerases at the eukaryotic fork-20 years later
Abstract
Function of the eukaryotic genome depends on efficient and accurate replication of anti-parallel DNA strands. Eukaryotic DNA polymerases have different properties adapted to perform a wide spectrum of DNA transactions. Here we focus on major players in the bulk replication, DNA polymerases of the B-family. We review the organization of the replication fork in eukaryotes in a historical perspective, analyze contemporary models and propose a new integrative model of the fork.
Copyright (c) 2009 Elsevier B.V. All rights reserved.
Figures
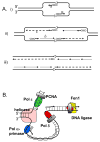

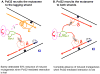
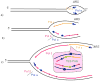
References
-
- Morrison A, Araki H, Clark AB, Hamatake RK, Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1990;62:1143–1151. - PubMed
-
- Chilkova O, Stenlund P, Isoz I, Stith CM, Grabowski P, Lundstrom EB, Burgers PM, Johansson E. The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNANucleic. Acids Res. 2007;35:6588–6597. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

