Identification of selective inhibitors of cancer stem cells by high-throughput screening
- PMID: 19682730
- PMCID: PMC4892125
- DOI: 10.1016/j.cell.2009.06.034
Identification of selective inhibitors of cancer stem cells by high-throughput screening
Abstract
Screens for agents that specifically kill epithelial cancer stem cells (CSCs) have not been possible due to the rarity of these cells within tumor cell populations and their relative instability in culture. We describe here an approach to screening for agents with epithelial CSC-specific toxicity. We implemented this method in a chemical screen and discovered compounds showing selective toxicity for breast CSCs. One compound, salinomycin, reduces the proportion of CSCs by >100-fold relative to paclitaxel, a commonly used breast cancer chemotherapeutic drug. Treatment of mice with salinomycin inhibits mammary tumor growth in vivo and induces increased epithelial differentiation of tumor cells. In addition, global gene expression analyses show that salinomycin treatment results in the loss of expression of breast CSC genes previously identified by analyses of breast tissues isolated directly from patients. This study demonstrates the ability to identify agents with specific toxicity for epithelial CSCs.
Figures
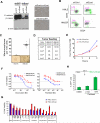
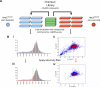
 ) denoting the CSC-enriched fraction and the blue ellipse (
) denoting the CSC-enriched fraction and the blue ellipse ( ) the CSC-depleted fraction. (b) Quantification of tumorsphere-formation with HMLER cells treated as in (a). (c) Heterogeneous populations (control/EMT mixtures) of HMLE and HMLER cells (HMLE_Mx, HMLER_Mx, respectively) were compound-treated for 4 days, cultured in the absence of compound for 4 days, and the percent of CD44high/CD24low cells quantified by FACS. (d) Quantification of mammosphere-formation in HMLE_Mx and HMLER_Mx populations compound-treated as in (a). Phase-contrast images of mammospheres are shown. (e)
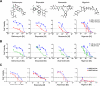
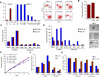
In vitro growth curves of HMLER cells compound-treated as in (a) are shown. (f) Compound-pretreated MCF7Ras (4000 cells/well) and 4T1 cells (1000 cells/well) were seeded in the absence of compound and tumorsphere formation assessed at 10 days. (g) The fraction of viable cells after compound treatment was assessed using trypan-blue exclusion for both the parental 4T1 line and a paclitaxel-resistant 4T1 line (4T1-TaxR).
) the CSC-depleted fraction. (b) Quantification of tumorsphere-formation with HMLER cells treated as in (a). (c) Heterogeneous populations (control/EMT mixtures) of HMLE and HMLER cells (HMLE_Mx, HMLER_Mx, respectively) were compound-treated for 4 days, cultured in the absence of compound for 4 days, and the percent of CD44high/CD24low cells quantified by FACS. (d) Quantification of mammosphere-formation in HMLE_Mx and HMLER_Mx populations compound-treated as in (a). Phase-contrast images of mammospheres are shown. (e)
In vitro growth curves of HMLER cells compound-treated as in (a) are shown. (f) Compound-pretreated MCF7Ras (4000 cells/well) and 4T1 cells (1000 cells/well) were seeded in the absence of compound and tumorsphere formation assessed at 10 days. (g) The fraction of viable cells after compound treatment was assessed using trypan-blue exclusion for both the parental 4T1 line and a paclitaxel-resistant 4T1 line (4T1-TaxR).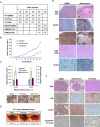

Comment in
-
Breast cancer stem cells: eradication by differentiation therapy?Cell. 2009 Aug 21;138(4):623-5. doi: 10.1016/j.cell.2009.08.007. Cell. 2009. PMID: 19703390
References
-
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. - PubMed
-
- Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nature Rev Cancer. 2005;5:275–284. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

