Transcriptional regulation of mitochondrial glycerophosphate acyltransferase is mediated by distal promoter via ChREBP and SREBP-1
- PMID: 19682972
- PMCID: PMC2761506
- DOI: 10.1016/j.abb.2009.07.027
Transcriptional regulation of mitochondrial glycerophosphate acyltransferase is mediated by distal promoter via ChREBP and SREBP-1
Abstract
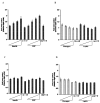
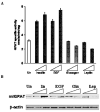
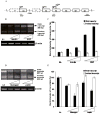
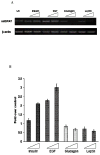
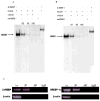
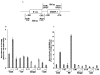
We have recently identified two promoters, distal and proximal for rat mitochondrial glycerophosphate acyltransferase (mtGPAT). Here we are reporting further characterization of the promoters. Insulin and epidermal growth factor (EGF) stimulated while leptin and glucagon inhibited the luciferase activity of the distal promoter and the amounts of the distal transcript. Conversely, luciferase activity of the proximal promoter and proximal transcript remained unchanged due to these treatments. Only the distal promoter has binding sites for carbohydrate response element binding protein (ChREBP) and sterol regulatory element binding protein-1 (SREBP-1). Electromobility shift assays and chromatin immunoprecipitation assays demonstrated that ChREBP and SREBP-1 bind to the mtGPAT distal promoter. Insulin and EGF increased while glucagon and leptin decreased the binding of SREBP-1 and ChREBP to the distal promoter. Thus, the distal promoter is the regulatory promoter while the proximal promoter acts constitutively for rat mtGPAT gene under the influence of hormones and growth factor.
Figures


References
-
- Haldar D, Kelker HC, Pullman ME. Properties of acyl coenzyme A: 1-acyl-sn-glycerol 3-phosphate acyltransferase from rat liver mitochondria and microsomes. Trans N Y Acad Sci. 1983;41:173–182. - PubMed
-
- Linden D, William-Olsson L, Rhedin M, Asztely AK, Clapham JC, Schreyer S. Overexpression of mitochondrial GPAT in rat hepatocytes leads to decreased fatty acid oxidation and increased glycerolipid biosynthesis. J Lipid Res. 2004;45:1279–1288. - PubMed
-
- Onorato TM, Chakraborty S, Balija VS, Haldar D. In: LIPIDS: Glycerolipid Metabolizing Enzymes. Haldar D, Das SK, editors. Research Signpost; Trivandrum: 2002.
-
- Hammond LE, Gallagher PA, Wang S, Hiller S, Kluckman KD, Posey-Marcos EL, Maeda N, Coleman RA. Mitochondrial glycerol-3-phosphate acyltransferase-deficient mice have reduced weight and liver triacylglycerol content and altered glycerolipid fatty acid composition. Mol Cell Biol. 2002;22:8204–8214. - PMC - PubMed
-
- Lindén D, William-Olsson L, Ahnmark A, Ekroos K, Hallberg C, Sjögren HP, Becker B, Svensson L, Clapham JC, Oscarsson J, Schreyer S. Liver-directed overexpression of mitochondrial glycerol-3-phosphate acyltransferase results in hepatic steatosis, increased triacylglycerol secretion and reduced fatty acid oxidation. FASEB J. 2006;20:434–443. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

