Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance
- PMID: 19683498
- PMCID: PMC2749492
- DOI: 10.1016/j.molcel.2009.06.015
Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance
Abstract
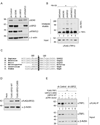
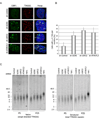
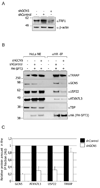
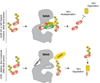
Histone acetyltransferases (HATs) play important roles in gene regulation and DNA repair by influencing the accessibility of chromatin to transcription factors and repair proteins. Here, we show that deletion of Gcn5 leads to telomere dysfunction in mouse and human cells. Biochemical studies reveal that depletion of Gcn5 or ubiquitin-specific protease 22 (Usp22), which is another bona fide component of the Gcn5-containing SAGA complex, increases ubiquitination and turnover of TRF1, a primary component of the telomeric shelterin complex. Inhibition of the proteasome or overexpression of USP22 opposes this effect. The USP22 deubiquitinating module requires association with SAGA complexes for activity, and we find that depletion of Gcn5 compromises this association in mammalian cells. Thus, our results indicate that Gcn5 regulates TRF1 levels through effects on Usp22 activity and SAGA integrity.
Figures



Comment in
-
The SAGA continues...to the end.Mol Cell. 2009 Aug 14;35(3):256-8. doi: 10.1016/j.molcel.2009.07.011. Mol Cell. 2009. PMID: 19683489
References
-
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. - PubMed
-
- Bonnet J, Romier C, Tora L, Devys D. Zinc-finger UBPs: regulators of deubiquitylation. Trends Biochem Sci. 2008;33:369–375. - PubMed
-
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. - PubMed
-
- Candau R, Berger SL. Structural and functional analysis of yeast putative adaptors. Evidence for an adaptor complex in vivo. J Biol Chem. 1996;271:5237–5245. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

