Human copper transporter 1 lacking O-linked glycosylation is proteolytically cleaved in a Rab9-positive endosomal compartment
- PMID: 19684018
- PMCID: PMC2788861
- DOI: 10.1074/jbc.M109.044925
Human copper transporter 1 lacking O-linked glycosylation is proteolytically cleaved in a Rab9-positive endosomal compartment
Abstract
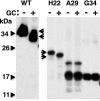
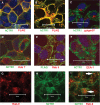
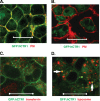
The human copper transporter hCTR1 is a homotrimer composed of a plasma membrane protein of 190 amino acids that contains three transmembrane segments. The extracellular 65-amino acid amino terminus of hCTR1 contains both N-linked (at Asn(15)) and O-linked (at Thr(27)) sites of glycosylation. If O-glycosylation at Thr(27) is prevented, hCTR1 is efficiently cleaved, removing approximately 30 amino acids from the amino terminus. We have now investigated (i) the site of this cleavage, determining which peptide bonds are cleaved, (ii) the mechanism by which glycosylation prevents cleavage, and (iii) where in the cell the proteolytic cleavage takes place. Cleavage occurs in the sequence Ala-Ser-His-Ser-His (residues 29-33), which does not contain previously recognized protease cleavage sites. Using a series of hCTR1 mutants, we show that cleavage occurs preferentially between residues Ala(29)-Ser(30)-His(31). We also show that the O-linked polysaccharide at Thr(27) blocks proteolysis due to its proximity to the cleavage site. Moving the cleavage site away from the Thr(27) polysaccharide by insertion of as few as 5 amino acids allows cleavage to occur in the presence of glycosylation. Imaging studies using immunofluorescence in fixed cells and a functional green fluorescent protein-tagged hCTR1 transporter in live cells showed that the cleaved peptide accumulates in punctate structures in the cytoplasm. These puncta overlap compartments were stained by Rab9, indicating that hCTR1 cleavage occurs in a late endosomal compartment prior to delivery of the transporter to the plasma membrane.
Figures







References
-
- Petris M. J. (2004) Pflugers Arch. 447, 752–755 - PubMed
-
- Linder M. C., Wooten L., Cerveza P., Cotton S., Shulze R., Lomeli N. (1998) Am. J. Clin. Nutr. 67, Suppl. 5, 965S–971S - PubMed
-
- Maryon E. B., Molloy S. A., Zimnicka A. M., Kaplan J. H. (2007) Biometals 20, 355–364 - PubMed
-
- Kim B. E., Nevitt T., Thiele D. J. (2008) Nat. Chem. Biol. 4, 176–185 - PubMed
-
- Huffman D. L., O'Halloran T. V. (2001) Annu. Rev. Biochem. 70, 677–701 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

