A Rab escort protein integrates the secretion system with TOR signaling and ribosome biogenesis
- PMID: 19684114
- PMCID: PMC2725937
- DOI: 10.1101/gad.1804409
A Rab escort protein integrates the secretion system with TOR signaling and ribosome biogenesis
Abstract
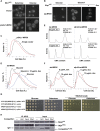
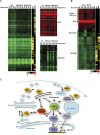
The coupling of environmental conditions to cell growth and division is integral to cell fitness. In Saccharomyces cerevisiae, the transcription factor Sfp1 couples nutrient status to cell growth rate by controlling the expression of ribosome biogenesis (Ribi) and ribosomal protein (RP) genes. Sfp1 is localized to the nucleus in rich nutrients, but upon nutrient limitation or target of rapamycin (TOR) pathway inhibition by rapamycin, Sfp1 rapidly exits the nucleus, leading to repression of the Ribi/RP regulons. Through systematic cell-based screens we found that many components of the secretory system influence Sfp1 localization. Notably, the essential Rab escort protein Mrs6 exhibited a nutrient-sensitive interaction with Sfp1. Overexpression of Mrs6 prevented nuclear localization of Sfp1 in rich nutrients, whereas loss of Mrs6 resulted in nuclear Sfp1 localization in poor nutrients. These effects were specific to Sfp1 and independent of the protein kinase C (PKC) pathway, suggesting that Mrs6 lies in a distinct branch of TOR and ribosome biogenesis regulation. Rapamycin-resistant alleles of MRS6 were defective in the cytoplasmic retention of Sfp1, the control of cell size, and in the repression of the Ribi/RP regulons. The Sfp1-Mrs6 interaction is a nexus for growth regulation that links the secretory system and TOR-dependent nutrient signaling to ribosome biogenesis.
Figures




Similar articles
-
Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling.Mol Cell. 2009 Mar 27;33(6):704-16. doi: 10.1016/j.molcel.2009.01.034. Mol Cell. 2009. PMID: 19328065
-
Proteasomal degradation of Sfp1 contributes to the repression of ribosome biogenesis during starvation and is mediated by the proteasome activator Blm10.Mol Biol Cell. 2011 Mar 1;22(5):528-40. doi: 10.1091/mbc.E10-04-0352. Epub 2011 Jan 5. Mol Biol Cell. 2011. PMID: 21209318 Free PMC article.
-
Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression.Proc Natl Acad Sci U S A. 2004 Oct 5;101(40):14315-22. doi: 10.1073/pnas.0405353101. Epub 2004 Sep 7. Proc Natl Acad Sci U S A. 2004. PMID: 15353587 Free PMC article.
-
Transcriptional control of ribosome biogenesis in yeast: links to growth and stress signals.Biochem Soc Trans. 2021 Aug 27;49(4):1589-1599. doi: 10.1042/BST20201136. Biochem Soc Trans. 2021. PMID: 34240738 Free PMC article. Review.
-
Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae.Microbiol Mol Biol Rev. 2002 Dec;66(4):579-91, table of contents. doi: 10.1128/MMBR.66.4.579-591.2002. Microbiol Mol Biol Rev. 2002. PMID: 12456783 Free PMC article. Review.
Cited by
-
Nutritional control of growth and development in yeast.Genetics. 2012 Sep;192(1):73-105. doi: 10.1534/genetics.111.135731. Genetics. 2012. PMID: 22964838 Free PMC article. Review.
-
REP1 inhibits FOXO3-mediated apoptosis to promote cancer cell survival.Cell Death Dis. 2017 Jan 5;8(1):e2536. doi: 10.1038/cddis.2016.462. Cell Death Dis. 2017. PMID: 28055019 Free PMC article.
-
A Rab escort protein regulates the MAPK pathway that controls filamentous growth in yeast.Sci Rep. 2020 Dec 17;10(1):22184. doi: 10.1038/s41598-020-78470-4. Sci Rep. 2020. PMID: 33335117 Free PMC article.
-
Nuclear envelope expansion in budding yeast is independent of cell growth and does not determine nuclear volume.Mol Biol Cell. 2019 Jan 1;30(1):131-145. doi: 10.1091/mbc.E18-04-0204. Epub 2018 Oct 31. Mol Biol Cell. 2019. PMID: 30379612 Free PMC article.
-
Growth-dependent signals drive an increase in early G1 cyclin concentration to link cell cycle entry with cell growth.Elife. 2021 Oct 29;10:e64364. doi: 10.7554/eLife.64364. Elife. 2021. PMID: 34713806 Free PMC article.
References
-
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. - PubMed
-
- Bialek-Wyrzykowska U, Bauer BE, Wagner W, Kohlwein SD, Schweyen RJ, Ragnini A. Low levels of Ypt protein prenylation cause vesicle polarization defects and thermosensitive growth that can be suppressed by genes involved in cell wall maintenance. Mol Microbiol. 2000;35:1295–1311. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
