Human papillomaviruses: genetic basis of carcinogenicity
- PMID: 19684441
- PMCID: PMC2835381
- DOI: 10.1159/000214919
Human papillomaviruses: genetic basis of carcinogenicity
Abstract
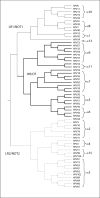
Persistent infection by specific oncogenic human papillomaviruses (HPVs) is established as the necessary cause of cervix cancer. DNA sequence differences between HPV genomes determine whether an HPV has the potential to cause cancer. Of the more than 100 HPV genotypes characterized at the genetic level, at least 15 are associated, to varying degrees, with cervical cancer. Classification based on nucleotide similarity places nearly all HPVs that infect the cervicovaginal area within the alpha-PV genus. Within this genus, phylogenetic trees inferred from the entire viral genome cluster all cancer-causing types together, suggesting the existence of a common ancestor for the oncogenic HPVs. However, in separate trees built from the early open reading frames (ORFs; i.e. E1, E2, E6, E7) or the late ORFs (i.e. L1, L2), the carcinogenic potential sorts with the early region of the genome, but not the late region. Thus, genetic differences within the early region specify the pathogenic potential of alpha-HPV infections. Since the HPV genomes are monophyletic and sites are highly correlated across the genome, diagnosis of oncogenic types and non-oncogenic types can be accomplished using any region across the genome. Here we review our current understanding of the evolutionary history of the oncogenic HPVs, in particular, we focus on the importance of viral genome heterogeneity and discuss the genetic basis for the oncogenic phenotype in some but not all alpha-PVs.
Copyright 2009 S. Karger AG, Basel.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

