Silencing by small RNAs is linked to endosomal trafficking
- PMID: 19684574
- PMCID: PMC2737091
- DOI: 10.1038/ncb1930
Silencing by small RNAs is linked to endosomal trafficking
Erratum in
- Nat Cell Biol. 2009 Dec;11(12):1495
Abstract
Small RNAs direct RNA-induced silencing complexes (RISCs) to regulate stability and translation of mRNAs. RISCs associated with target mRNAs often accumulate in discrete cytoplasmic foci known as GW-bodies. However, RISC proteins can associate with membrane compartments such as the Golgi and endoplasmic reticulum. Here, we show that GW-bodies are associated with late endosomes (multivesicular bodies, MVBs). Blocking the maturation of MVBs into lysosomes by loss of the tethering factor HPS4 (ref. 5) enhances short interfering RNA (siRNA)- and micro RNA (miRNA)-mediated silencing in Drosophila melanogaster and humans. It also triggers over-accumulation of GW-bodies. Blocking MVB formation by ESCRT (endosomal sorting complex required for transport) depletion results in impaired miRNA silencing and loss of GW-bodies. These results indicate that active RISCs are physically and functionally coupled to MVBs. We further show that MVBs promote the competence of RISCs in loading small RNAs. We suggest that the recycling of RISCs is promoted by MVBs, resulting in RISCs more effectively engaging with small RNA effectors and possibly target RNAs. It may provide a means to enhance the dynamics of RNA silencing in the cytoplasm.
Figures
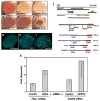
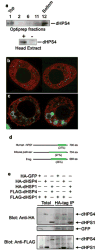

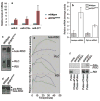

Comment in
-
Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity.Nat Cell Biol. 2009 Sep;11(9):1143-9. doi: 10.1038/ncb1929. Epub 2009 Aug 16. Nat Cell Biol. 2009. PMID: 19684575
-
RISC hitches onto endosome trafficking.Nat Cell Biol. 2009 Sep;11(9):1049-51. doi: 10.1038/ncb0909-1049. Nat Cell Biol. 2009. PMID: 19724258
References
-
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–24. - PubMed
-
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. - PubMed
-
- Li W, et al. Murine Hermansky-Pudlak syndrome genes: regulators of lysosome-related organelles. Bioessays. 2004;26:616–28. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

