A 3D digital atlas of C. elegans and its application to single-cell analyses
- PMID: 19684595
- PMCID: PMC2882208
- DOI: 10.1038/nmeth.1366
A 3D digital atlas of C. elegans and its application to single-cell analyses
Abstract
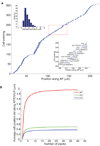
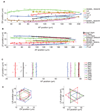
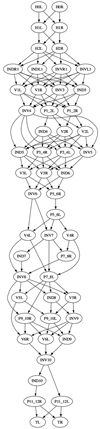
We built a digital nuclear atlas of the newly hatched, first larval stage (L1) of the wild-type hermaphrodite of Caenorhabditis elegans at single-cell resolution from confocal image stacks of 15 individual worms. The atlas quantifies the stereotypy of nuclear locations and provides other statistics on the spatial patterns of the 357 nuclei that could be faithfully segmented and annotated out of the 558 present at this developmental stage. We then developed an automated approach to assign cell names to each nucleus in a three-dimensional image of an L1 worm. We achieved 86% accuracy in identifying the 357 nuclei automatically. This computational method will allow high-throughput single-cell analyses of the post-embryonic worm, such as gene expression analysis, or ablation or stimulation of cells under computer control in a high-throughput functional screen.
Figures


Comment in
-
Google 'EarthWorm'.Nat Methods. 2009 Sep;6(9):635-6. doi: 10.1038/nmeth0909-635. Nat Methods. 2009. PMID: 19718015 No abstract available.
References
-
- Riddle DL, Blumenthal T, Meyer BJ, Priess JR. C. elegans II. Cold Sprint Harbor Laboratory Press; 1997. - PubMed
-
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. - PubMed
-
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of he nematode, Caenorhabditis elegmans. Dev. Biol. 1977;56:110–156. - PubMed
-
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode C. elegans. Phil. Trans. Royal Soc. London. Series B, Biol Scien. 1986;314:1. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

