A gene expression signature classifying telomerase and ALT immortalization reveals an hTERT regulatory network and suggests a mesenchymal stem cell origin for ALT
- PMID: 19684619
- PMCID: PMC2875172
- DOI: 10.1038/onc.2009.238
A gene expression signature classifying telomerase and ALT immortalization reveals an hTERT regulatory network and suggests a mesenchymal stem cell origin for ALT
Abstract
Telomere length is maintained by two known mechanisms, the activation of telomerase or alternative lengthening of telomeres (ALT). The molecular mechanisms regulating the ALT phenotype are poorly understood and it is unknown how the decision of which pathway to activate is made at the cellular level. We have shown earlier that active repression of telomerase gene expression by chromatin remodelling of the promoters is one mechanism of regulation; however, other genes and signalling networks are likely to be required to regulate telomerase and maintain the ALT phenotype. Using gene expression profiling, we have uncovered a signature of 1305 genes to distinguish telomerase-positive and ALT cell lines. By combining this with the gene expression profiles of liposarcoma tissue samples, we refined this signature to 297 genes. A network analysis of known interactions between genes within this signature revealed a regulatory signalling network consistent with a model of human telomerase reverse transcriptase (hTERT) repression in ALT cell lines and liposarcomas. This network expands on our existing knowledge of hTERT regulation and provides a platform to understand differential regulation of hTERT in different tumour types and normal tissues. We also show evidence to suggest a novel mesenchymal stem cell origin for ALT immortalization in cell lines and mesenchymal tissues.
Figures



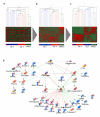

References
-
- Atkinson SP, Hoare SF, Glasspool RM, Keith WN. Lack of telomerase gene expression in alternative lengthening of telomere cells is associated with chromatin remodeling of the hTR and hTERT gene promoters. Cancer Res. 2005;65:7585–7590. - PubMed
-
- Bernardo ME, Zaffaroni N, Novara F, Cometa AM, Avanzini MA, Moretta A, et al. Human bone marrow derived mesenchymal stem cells do not undergo transformation after long-term in vitro culture and do not exhibit telomere maintenance mechanisms. Cancer Res. 2007;67:9142–9149. - PubMed
-
- Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–1274. - PubMed
-
- Cairney CJ, Keith WN. Telomerase redefined: integrated regulation of hTR and hTERT for telomere maintenance and telomerase activity. Biochimie. 2008;90:13–23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

