Reproducibility of Genotypes as Measured by the Affymetrix GeneChip® 100K Human Mapping Array Set
- PMID: 19684844
- PMCID: PMC2597860
- DOI: 10.1016/j.csda.2008.05.020
Reproducibility of Genotypes as Measured by the Affymetrix GeneChip® 100K Human Mapping Array Set
Abstract
Genotyping errors that are undetected in genome-wide association studies using single nucleotide polymorphisms (SNPs) may degrade the likelihood of detecting true positive associations. To estimate the frequency of genotyping errors and assess the reproducibility of genotype calls, we analyzed two sets of duplicate data, one dataset containing twenty blind duplicates and another dataset containing twenty-eight non-random duplicates, from a genome-wide association study using Affymetrix GeneChip® 100K Human Mapping Arrays. For the twenty blind duplicates the overall agreement in genotyping calls as measured with the Kappa statistics, was 0.997, with a discordancy rate of 0.27%. For the twenty-eight nonrandom duplicates, the overall agreement was lower, 0.95, with a higher discordancy rate of 4.53%. The accuracy and probability of concordancy were inversely related to the genotyping uncertainty score, i.e., as the genotyping uncertainty score increased, the concordancy and probability of concordant calls decreased. Lowering of the uncertainty score threshold for rejection of genotype calls from the Affymetrix recommended value of 0.25 to 0.20 resulted in an increased predicted accuracy from 92.6% to 95% with a slight increase in the "No Call" rate from 1.81% to 2.33%. Hence, we suggest using a lower uncertainty score threshold, say 0.20, which will result in higher accuracy in calls at a modest decrease in the call rate.
Figures

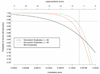
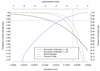
References
-
- Ahn K, Haynes C, Kim W, Fleur RS, Gordon D, Finch SJ. The effects of SNP genotyping errors on the power of the Cochran-Armitage linear trend test for case/control association studies. Ann Hum Genet. 2007;71:249–261. - PubMed
-
- Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P. How to track and assess genotyping errors in population genetics studies. Molecular Ecology. 2004;13:3261–3273. - PubMed
-
- Chapman AB, Schwartz GL, Boerwinkle E, Turner ST. Predictors of antihypertensive response to a standard dose of hydrochlorothiazide for essential hypertension. Kidney International. 2002;61:1047–1055. - PubMed
-
- Collins FS, Guyer MS, Charkravarti A. Variations on a theme: cataloging human DNA sequence variation. Science. 1997;278:1580–1581. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
