Parallel expression evolution of oxidative stress-related genes in fiber from wild and domesticated diploid and polyploid cotton (Gossypium)
- PMID: 19686594
- PMCID: PMC2907704
- DOI: 10.1186/1471-2164-10-378
Parallel expression evolution of oxidative stress-related genes in fiber from wild and domesticated diploid and polyploid cotton (Gossypium)
Abstract
Background: Reactive oxygen species (ROS) play a prominent role in signal transduction and cellular homeostasis in plants. However, imbalances between generation and elimination of ROS can give rise to oxidative stress in growing cells. Because ROS are important to cell growth, ROS modulation could be responsive to natural or human-mediated selection pressure in plants. To study the evolution of oxidative stress related genes in a single plant cell, we conducted comparative expression profiling analyses of the elongated seed trichomes ("fibers") of cotton (Gossypium), using a phylogenetic approach.
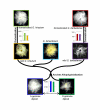
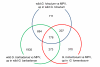
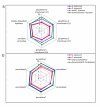
Results: We measured expression changes during diploid progenitor species divergence, allopolyploid formation and parallel domestication of diploid and allopolyploid species, using a microarray platform that interrogates 42,429 unigenes. The distribution of differentially expressed genes in progenitor diploid species revealed significant up-regulation of ROS scavenging and potential signaling processes in domesticated G. arboreum. Similarly, in two independently domesticated allopolyploid species (G. barbadense and G. hirsutum) antioxidant genes were substantially up-regulated in comparison to antecedent wild forms. In contrast, analyses of three wild allopolyploid species indicate that genomic merger and ancient allopolyploid formation had no significant influences on regulation of ROS related genes. Remarkably, many of the ROS-related processes diagnosed as possible targets of selection were shared among diploid and allopolyploid cultigens, but involved different sets of antioxidant genes.
Conclusion: Our data suggests that parallel human selection for enhanced fiber growth in several geographically widely dispersed species of domesticated cotton resulted in similar and overlapping metabolic transformations of the manner in which cellular redox levels have become modulated.
Figures

Similar articles
-
Global expression dynamics and miRNA evolution profile govern floral/fiber architecture in the modern cotton (Gossypium).Planta. 2021 Aug 30;254(3):62. doi: 10.1007/s00425-021-03711-3. Planta. 2021. PMID: 34459999
-
Independent Domestication of Two Old World Cotton Species.Genome Biol Evol. 2016 Jul 2;8(6):1940-7. doi: 10.1093/gbe/evw129. Genome Biol Evol. 2016. PMID: 27289095 Free PMC article.
-
Proteomic profiling of developing cotton fibers from wild and domesticated Gossypium barbadense.New Phytol. 2013 Oct;200(2):570-582. doi: 10.1111/nph.12381. Epub 2013 Jun 25. New Phytol. 2013. PMID: 23795774
-
Parental squabbles and genome expression: lessons from the polyploids.J Biol. 2009;8(4):43. doi: 10.1186/jbiol140. Epub 2009 May 1. J Biol. 2009. PMID: 19439050 Free PMC article. Review.
-
An Overview of Cotton Gland Development and Its Transcriptional Regulation.Int J Mol Sci. 2022 Apr 28;23(9):4892. doi: 10.3390/ijms23094892. Int J Mol Sci. 2022. PMID: 35563290 Free PMC article. Review.
Cited by
-
Genetic Basis of Fiber Improvement and Decreased Stress Tolerance in Cultivated Versus Semi-Domesticated Upland Cotton.Front Plant Sci. 2019 Nov 29;10:1572. doi: 10.3389/fpls.2019.01572. eCollection 2019. Front Plant Sci. 2019. PMID: 31850042 Free PMC article.
-
GhABF2, a bZIP transcription factor, confers drought and salinity tolerance in cotton (Gossypium hirsutum L.).Sci Rep. 2016 Oct 7;6:35040. doi: 10.1038/srep35040. Sci Rep. 2016. PMID: 27713524 Free PMC article.
-
Conservation and Divergence in Duplicated Fiber Coexpression Networks Accompanying Domestication of the Polyploid Gossypium hirsutum L.G3 (Bethesda). 2020 Aug 5;10(8):2879-2892. doi: 10.1534/g3.120.401362. G3 (Bethesda). 2020. PMID: 32586849 Free PMC article.
-
Gene expression in developing fibres of Upland cotton (Gossypium hirsutum L.) was massively altered by domestication.BMC Biol. 2010 Nov 15;8:139. doi: 10.1186/1741-7007-8-139. BMC Biol. 2010. PMID: 21078138 Free PMC article.
-
SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs.PLoS Genet. 2011 May;7(5):e1001388. doi: 10.1371/journal.pgen.1001388. Epub 2011 May 26. PLoS Genet. 2011. PMID: 21637781 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

