The N-Myc-DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain
- PMID: 19686682
- PMCID: PMC2769073
- DOI: 10.1016/j.devcel.2009.07.009
The N-Myc-DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain
Abstract
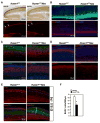
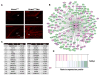
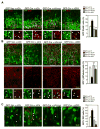
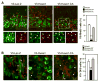
Self-renewal and proliferation of neural stem cells and the decision to initiate neurogenesis are crucial events directing brain development. Here we show that the ubiquitin ligase Huwe1 operates upstream of the N-Myc-DLL3-Notch pathway to control neural stem cell activity and promote neurogenesis. Conditional inactivation of the Huwe1 gene in the mouse brain caused neonatal lethality associated with disorganization of the laminar patterning of the cortex. These defects stemmed from severe impairment of neurogenesis associated with uncontrolled expansion of the neural stem cell compartment. Loss- and gain-of-function experiments in the mouse cortex demonstrated that Huwe1 restrains proliferation and enables neuronal differentiation by suppressing the N-Myc-DLL3 cascade. Notably, human high-grade gliomas carry focal hemizygous deletions of the X-linked Huwe1 gene in association with amplification of the N-myc locus. Our results indicate that Huwe1 balances proliferation and neurogenesis in the developing brain and that this pathway is subverted in malignant brain tumors.
Figures



References
-
- Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. - PubMed
-
- Bahram F, von der Lehr N, Cetinkaya C, Larsson LG. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood. 2000;95:2104–2110. - PubMed
-
- Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. - PubMed
-
- Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21. - PubMed
-
- Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt's lymphoma and mouse plasmacytomas. Nat Genet. 1993;5:56–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

