The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1
- PMID: 19686687
- PMCID: PMC2908246
- DOI: 10.1016/j.devcel.2009.06.007
The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1
Abstract
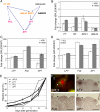
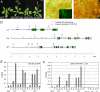
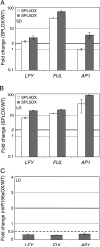
When to form flowers is a developmental decision that profoundly impacts the fitness of flowering plants. In Arabidopsis this decision is ultimately controlled by the induction and subsequent activity of the transcription factors LEAFY (LFY), FRUITFULL (FUL), and APETALA1 (AP1). Despite their central importance, our current understanding of the regulation of LFY, FUL, and AP1 expression is still incomplete. We show here that all three genes are directly activated by the microRNA-targeted transcription factor SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 (SPL3). Our findings suggest that SPL3 acts together with other microRNA-regulated SPL transcription factors to control the timing of flower formation. Moreover, the identified SPL activity defines a distinct pathway in control of this vital developmental decision.
Figures


References
-
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. - PubMed
-
- Achard P, Herr A, Baulcombe DC, Harberd NP. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131:3357–3365. - PubMed
-
- Araki T. Transition from vegetative to reproductive phase. Curr Opin Plant Biol. 2001;4:63–68. - PubMed
-
- Blazquez MA, Ferrandiz C, Madueno F, Parcy F. How floral meristems are built. Plant Mol Biol. 2006;60:855–870. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

