Alpha-defensins in enteric innate immunity: functional Paneth cell alpha-defensins in mouse colonic lumen
- PMID: 19687006
- PMCID: PMC2788835
- DOI: 10.1074/jbc.M109.050773
Alpha-defensins in enteric innate immunity: functional Paneth cell alpha-defensins in mouse colonic lumen
Abstract
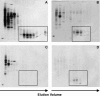
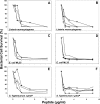
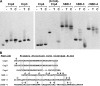
Paneth cells are a secretory epithelial lineage that release dense core granules rich in host defense peptides and proteins from the base of small intestinal crypts. Enteric alpha-defensins, termed cryptdins (Crps) in mice, are highly abundant in Paneth cell secretions and inherently resistant to proteolysis. Accordingly, we tested the hypothesis that enteric alpha-defensins of Paneth cell origin persist in a functional state in the mouse large bowel lumen. To test this idea, putative Crps purified from mouse distal colonic lumen were characterized biochemically and assayed in vitro for bactericidal peptide activities. The peptides comigrated with cryptdin control peptides in acid-urea-PAGE and SDS-PAGE, providing identification as putative Crps. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry experiments showed that the molecular masses of the putative alpha-defensins matched those of the six most abundant known Crps, as well as N-terminally truncated forms of each, and that the peptides contain six Cys residues, consistent with identities as alpha-defensins. N-terminal sequencing definitively revealed peptides with N termini corresponding to full-length, (des-Leu)-truncated, and (des-Leu-Arg)-truncated N termini of Crps 1-4 and 6. Crps from mouse large bowel lumen were bactericidal in the low micromolar range. Thus, Paneth cell alpha-defensins secreted into the small intestinal lumen persist as intact and functional forms throughout the intestinal tract, suggesting that the peptides may mediate enteric innate immunity in the colonic lumen, far from their upstream point of secretion in small intestinal crypts.
Figures


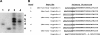
Similar articles
-
Activation of Paneth cell alpha-defensins in mouse small intestine.J Biol Chem. 2002 Feb 15;277(7):5219-28. doi: 10.1074/jbc.M109410200. Epub 2001 Dec 3. J Biol Chem. 2002. PMID: 11733520
-
Characterization of luminal paneth cell alpha-defensins in mouse small intestine. Attenuated antimicrobial activities of peptides with truncated amino termini.J Biol Chem. 2000 Oct 27;275(43):33969-73. doi: 10.1074/jbc.M004062200. J Biol Chem. 2000. PMID: 10942762
-
Paneth cell alpha-defensins from rhesus macaque small intestine.Infect Immun. 2004 Mar;72(3):1470-8. doi: 10.1128/IAI.72.3.1470-1478.2004. Infect Immun. 2004. PMID: 14977952 Free PMC article.
-
Paneth cell alpha-defensins: peptide mediators of innate immunity in the small intestine.Springer Semin Immunopathol. 2005 Sep;27(2):133-46. doi: 10.1007/s00281-005-0202-x. Epub 2005 Jun 2. Springer Semin Immunopathol. 2005. PMID: 15931529 Review.
-
Alpha-defensins in the gastrointestinal tract.Mol Immunol. 2003 Nov;40(7):463-7. doi: 10.1016/s0161-5890(03)00157-3. Mol Immunol. 2003. PMID: 14568393 Review.
Cited by
-
Elevated expression of Paneth cell CRS4C in ileitis-prone SAMP1/YitFc mice: regional distribution, subcellular localization, and mechanism of action.J Biol Chem. 2010 Mar 5;285(10):7493-504. doi: 10.1074/jbc.M109.083220. Epub 2010 Jan 7. J Biol Chem. 2010. PMID: 20056603 Free PMC article.
-
In vitro and in vivo synergistic effects of cryptdin 2 and ampicillin against Salmonella.Antimicrob Agents Chemother. 2011 Sep;55(9):4176-82. doi: 10.1128/AAC.00273-11. Epub 2011 Jun 20. Antimicrob Agents Chemother. 2011. PMID: 21690282 Free PMC article.
-
Recipient single nucleotide polymorphisms in Paneth cell antimicrobial peptide genes and acute graft-versus-host disease: analysis of BMT CTN-0201 and -0901 samples.Br J Haematol. 2018 Sep;182(6):887-894. doi: 10.1111/bjh.15492. Epub 2018 Jul 13. Br J Haematol. 2018. PMID: 30004111 Free PMC article. Clinical Trial.
-
Inflammatory bowel disease: an impaired barrier disease.Langenbecks Arch Surg. 2013 Jan;398(1):1-12. doi: 10.1007/s00423-012-1030-9. Epub 2012 Nov 18. Langenbecks Arch Surg. 2013. PMID: 23160753 Review.
-
Alpha-defensin 5 differentially modulates adenovirus vaccine vectors from different serotypes in vivo.PLoS Pathog. 2019 Dec 16;15(12):e1008180. doi: 10.1371/journal.ppat.1008180. eCollection 2019 Dec. PLoS Pathog. 2019. PMID: 31841560 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

