Elucidation of the ELK1 target gene network reveals a role in the coordinate regulation of core components of the gene regulation machinery
- PMID: 19687146
- PMCID: PMC2775591
- DOI: 10.1101/gr.093047.109
Elucidation of the ELK1 target gene network reveals a role in the coordinate regulation of core components of the gene regulation machinery
Abstract
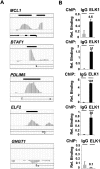
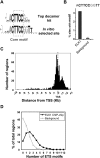
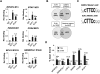
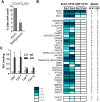
Transcription factors play an important role in orchestrating the activation of specific networks of genes through targeting their proximal promoter and distal enhancer regions. However, it is unclear how the specificity of downstream responses is maintained by individual members of transcription-factor families and, in most cases, what their target repertoire is. We have used ChIP-chip analysis to identify the target genes of the ETS-domain transcription factor ELK1. Two distinct modes of ELK1 target gene selection are identified; the first involves redundant promoter binding with other ETS-domain family members; the second occurs through combinatorial binding with a second transcription factor SRF, which specifies a unique group of target genes. One of the most prominent groups of genes forming the ELK1 target network includes classes involved in core gene expression control, namely, components of the basal transcriptional machinery, the spliceosome and the ribosome. Amongst the set of genes encoding the basal transcription machinery components, are a functionally linked subset of GTFs and TAFs. Our study, therefore, reveals an unsuspected level of coordinate regulation of components of the core gene expression control machinery and also identifies two different modes of promoter targeting through binding with a second transcription factor or redundant binding with other ETS-domain family members.
Figures


References
-
- Albright SR, Tjian R. TAFs revisited: More data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. - PubMed
-
- Bohlander SK, Espinosa R, III, Le Beau MM, Rowley JD, Díaz MO. A method for the rapid sequence-independent amplification of microdissected chromosomal material. Genomics. 1992;13:1322–1324. - PubMed
-
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. - PubMed
-
- Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
