Myosin-driven peroxisome partitioning in S. cerevisiae
- PMID: 19687257
- PMCID: PMC2733749
- DOI: 10.1083/jcb.200904050
Myosin-driven peroxisome partitioning in S. cerevisiae
Abstract
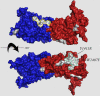
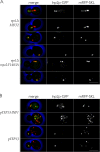
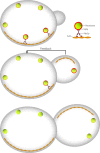
In Saccharomyces cerevisiae, the class V myosin motor Myo2p propels the movement of most organelles. We recently identified Inp2p as the peroxisome-specific receptor for Myo2p. In this study, we delineate the region of Myo2p devoted to binding peroxisomes. Using mutants of Myo2p specifically impaired in peroxisome binding, we dissect cell cycle-dependent and peroxisome partitioning-dependent mechanisms of Inp2p regulation. We find that although total Inp2p levels oscillate with the cell cycle, Inp2p levels on individual peroxisomes are controlled by peroxisome inheritance, as Inp2p aberrantly accumulates and decorates all peroxisomes in mother cells when peroxisome partitioning is abolished. We also find that Inp2p is a phosphoprotein whose level of phosphorylation is coupled to the cell cycle irrespective of peroxisome positioning in the cell. Our findings demonstrate that both organelle positioning and cell cycle progression control the levels of organelle-specific receptors for molecular motors to ultimately achieve an equidistribution of compartments between mother and daughter cells.
Figures




References
-
- Arai S., Noda Y., Kainuma S., Wada I., Yoda K. 2008. Ypt11 functions in bud-directed transport of the Golgi by linking Myo2 to the coatomer subunit Ret2.Curr. Biol. 18:987–991 - PubMed
-
- Beach D.L., Thibodeaux J., Maddox P., Yeh E., Bloom K. 2000. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast.Curr. Biol. 10:1497–1506 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

