FoxA1 binding directs chromatin structure and the functional response of a glucocorticoid receptor-regulated promoter
- PMID: 19687299
- PMCID: PMC2756877
- DOI: 10.1128/MCB.00368-09
FoxA1 binding directs chromatin structure and the functional response of a glucocorticoid receptor-regulated promoter
Abstract
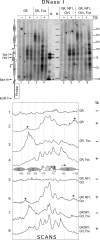
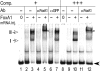
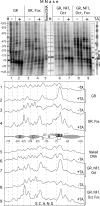
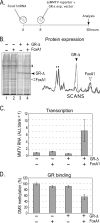
Reconstitution of the glucocorticoid receptor (GR)-regulated mouse mammary tumor virus (MMTV) promoter in Xenopus oocytes was used to monitor the effects of different transcription factor contexts. Three constitutively binding factors, nuclear factor 1 (NF1), octamer transcription factor 1 (Oct1), and the Forkhead box A1 (FoxA1), were shown to act in concert, to direct the chromatin structure, and to enhance the GR response. FoxA1 has a dominant effect in the absence of hormone and induces a cluster of DNase I-hypersensitive sites in the segment comprising bp -400 to +25. This FoxA1-mediated chromatin remodeling does not induce MMTV transcription, as opposed to that of the GR. However, the robust FoxA1-dependent chromatin opening has the following drastic functional consequences on the hormone regulation: (i) GR-DNA binding is facilitated, as revealed by dimethyl sulfate in vivo footprinting, leading to increased hormone-induced transcription, and (ii) the GR antagonist RU486 is converted into a partial agonist in the presence of FoxA1 via ligand-independent GR activation. We conclude that FoxA1 mediates a preset chromatin structure and directs a context-specific response of a nuclear receptor. Furthermore, the alternative nucleosome arrangement induced by GR and FoxA1 implies this to be determined by constitutive binding of transcription factors rather than by the DNA sequence itself.
Figures




References
-
- Astrand, C., S. Belikov, and O. Wrange. 2009. Histone acetylation characterizes chromatin presetting by NF1 and Oct1 and enhances glucocorticoid receptor binding to the MMTV promoter. Exp. Cell Res. 315:2604-2615. - PubMed
-
- Badis, G., E. T. Chan, H. van Bakel, L. Pena-Castillo, D. Tillo, K. Tsui, C. D. Carlson, A. J. Gossett, M. J. Hasinoff, C. L. Warren, M. Gebbia, S. Talukder, A. Yang, S. Mnaimneh, D. Terterov, D. Coburn, A. Li Yeo, Z. X. Yeo, N. D. Clarke, J. D. Lieb, A. Z. Ansari, C. Nislow, and T. R. Hughes. 2008. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell 32:878-887. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
