Prognostic significance of copy-number alterations in multiple myeloma
- PMID: 19687334
- PMCID: PMC2754906
- DOI: 10.1200/JCO.2008.20.6136
Prognostic significance of copy-number alterations in multiple myeloma
Abstract
Purpose: Chromosomal aberrations are a hallmark of multiple myeloma but their global prognostic impact is largely unknown.
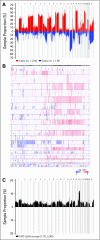
Patients and methods: We performed a genome-wide analysis of malignant plasma cells from 192 newly diagnosed patients with myeloma using high-density, single-nucleotide polymorphism (SNP) arrays to identify genetic lesions associated with prognosis.
Results: Our analyses revealed deletions and amplifications in 98% of patients. Amplifications in 1q and deletions in 1p, 12p, 14q, 16q, and 22q were the most frequent lesions associated with adverse prognosis, whereas recurrent amplifications of chromosomes 5, 9, 11, 15, and 19 conferred a favorable prognosis. Multivariate analysis retained three independent lesions: amp(1q23.3), amp(5q31.3), and del(12p13.31). When adjusted to the established prognostic variables (ie, t(4;14), del(17p), and serum beta(2)-microglobulin [Sbeta(2)M]), del(12p13.31) remained the most powerful independent adverse marker (P < .0001; hazard ratio [HR], 3.17) followed by Sbeta(2)M (P < .0001; HR, 2.78) and the favorable marker amp(5q31.3) (P = .0005; HR, 0.37). Patients with amp(5q31.3) alone and low Sbeta(2)M had an excellent prognosis (5-year overall survival, 87%); conversely, patients with del(12p13.31) alone or amp(5q31.3) and del(12p13.31) and high Sbeta(2)M had a very poor outcome (5-year overall survival, 20%). This prognostic model was validated in an independent validation cohort of 273 patients with myeloma.
Conclusion: These findings demonstrate the power and accessibility of molecular karyotyping to predict outcome in myeloma. In addition, integration of expression of genes residing in the lesions of interest revealed putative features of the disease driving short survival.
Conflict of interest statement
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Figures


Comment in
-
Heterogeneity in the prognostic significance of 12p deletion and chromosome 5 amplification in multiple myeloma.J Clin Oncol. 2011 Jan 10;29(2):e37-9; author reply e40-1. doi: 10.1200/JCO.2010.31.0516. Epub 2010 Dec 6. J Clin Oncol. 2011. PMID: 21135272 No abstract available.
References
-
- Smadja NV, Bastard C, Brigaudeau C, et al. Hypodiploidy is a major prognostic factorin multiple myeloma. Blood. 2001;98:2229–2238. - PubMed
-
- Fonseca R, Debes-Marun CS, Picken EB, et al. The recurrent IgH translocations are highly associated with nonhyperdiploid variant multiple myeloma. Blood. 2003;102:2562–2567. - PubMed
-
- Chng WJ, Kumar S, Vanwier S, et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res. 2007;67:2982–2989. - PubMed
-
- Zojer N, Königsberg R, Ackermann J, et al. Deletion of 13q14 remains an independent adverse prognostic variable in multiple myeloma despite its frequent detection by interphase fluorescence in situ hybridization. Blood. 2000;95:1925–1930. - PubMed
-
- Facon T, Avet-Loiseau H, Guillerm G, et al. Chromosome 13 abnormalities identified by FISH analysis and serum β2-microglobulin produce a very powerful myeloma staging system for patients receiving high dose therapy. Blood. 2001;97:1566–1571. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

