Residues in the H+ translocation site define the pKa for sugar binding to LacY
- PMID: 19689129
- PMCID: PMC2769999
- DOI: 10.1021/bi9011918
Residues in the H+ translocation site define the pKa for sugar binding to LacY
Abstract
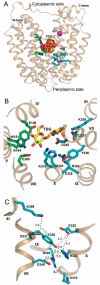
A remarkably high pKa of approximately 10.5 has been determined for sugar-binding affinity to the lactose permease of Escherichia coli (LacY), indicating that, under physiological conditions, substrate binds to fully protonated LacY. We have now systematically tested site-directed replacements for the residues involved in sugar binding, as well as H+ translocation and coupling, in order to determine which residues may be responsible for this alkaline pKa. Mutations in the sugar-binding site (Glu126, Trp151, Glu269) markedly decrease affinity for sugar but do not alter the pKa for binding. In contrast, replacements for residues involved in H+ translocation (Arg302, Tyr236, His322, Asp240, Glu325, Lys319) exhibit pKa values for sugar binding that are either shifted toward neutral pH or independent of pH. Values for the apparent dissociation constant for sugar binding (K(d)(app)) increase greatly for all mutants except neutral replacements for Glu325 or Lys319, which are characterized by remarkably high affinity sugar binding (i.e., low K(d)(app)) from pH 5.5 to pH 11. The pH dependence of the on- and off-rate constants for sugar binding measured directly by stopped-flow fluorometry implicates k(off) as a major factor for the affinity change at alkaline pH and confirms the effects of pH on K(d)(app) inferred from steady-state fluorometry. These results indicate that the high pKa for sugar binding by wild-type LacY cannot be ascribed to any single amino acid residue but appears to reside within a complex of residues involved in H+ translocation. There is structural evidence for water bound in this complex, and the water could be the site of protonation responsible for the pH dependence of sugar binding.
Figures









References
-
- Saier MH, Jr., Beatty JT, Goffeau A, Harley KT, Heijne WH, Huang SC, Jack DL, Jahn PS, Lew K, Liu J, Pao SS, Paulsen IT, Tseng TT, Virk PS. The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1999;1:257–279. - PubMed
-
- Saier MH., Jr. Families of transmembrane sugar transport proteins. Mol. Microbiol. 2000;35:699–710. - PubMed
-
- West IC. Lactose transport coupled to proton movements in Escherichia coli. Biochem. Biophys. Res. Commun. 1970;41:655–661. - PubMed
-
- Foster DL, Garcia ML, Newman MJ, Patel L, Kaback HR. Lactose-proton symport by purified lac carrier protein. Biochemistry. 1982;21:5634–5638. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

