Evolutionary dynamics of the LTR retrotransposons roo and rooA inferred from twelve complete Drosophila genomes
- PMID: 19689787
- PMCID: PMC3087523
- DOI: 10.1186/1471-2148-9-205
Evolutionary dynamics of the LTR retrotransposons roo and rooA inferred from twelve complete Drosophila genomes
Abstract
Background: Roo is the most abundant retrotransposon in the fruit fly Drosophila melanogaster. Its evolutionary origins and dynamics are thus of special interest for understanding the evolutionary history of Drosophila genome organization. We here study the phylogenetic distribution and evolution of roo, and its highly diverged relative rooA in 12 completely sequenced genomes of the genus Drosophila.
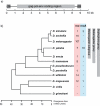
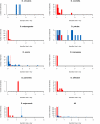
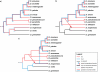
Results: We identify a total of 164 roo copies, 57 of which were previously unidentified copies that occur in 9 of the 12 genomes. Additionally we find 66 rooA copies in four genomes and remnants of this element in two additional genomes. We further increased the number of elements by searching for individual roo/rooA sequence domains. Most of our roo and rooA elements have been recently inserted. Most elements within a genome are highly similar. A comparison of the phylogenetic tree of our roo and rooA elements shows that the split between roo and rooA took place early in Drosophila evolution. Furthermore there is one incongruency between the species tree and the phylogenetic tree of the roo element. This incongruency regards the placement of elements from D. mojavensis, which are more closely related to D. melanogaster than elements from D. willistoni.
Conclusion: Within genomes, the evolutionary dynamics of roo and rooA range from recent transpositional activity to slow decay and extinction. Among genomes, the balance of phylogenetic evidence, sequence divergence distribution, and the occurrence of solo-LTR elements suggests an origin of roo/rooA within the Drosophila clade. We discuss the possibility of a horizontal gene transfer of roo within this clade.
Figures

Similar articles
-
Different structural variants of roo retrotransposon are active in Drosophila melanogaster.Gene. 2020 May 30;741:144546. doi: 10.1016/j.gene.2020.144546. Epub 2020 Mar 9. Gene. 2020. PMID: 32165306
-
The evolutionary dynamics of the Helena retrotransposon revealed by sequenced Drosophila genomes.BMC Evol Biol. 2009 Jul 22;9:174. doi: 10.1186/1471-2148-9-174. BMC Evol Biol. 2009. PMID: 19624823 Free PMC article.
-
Abundant and species-specific DINE-1 transposable elements in 12 Drosophila genomes.Genome Biol. 2008;9(2):R39. doi: 10.1186/gb-2008-9-2-r39. Epub 2008 Feb 21. Genome Biol. 2008. PMID: 18291035 Free PMC article.
-
Evolutionary forces generating sequence homogeneity and heterogeneity within retrotransposon families.Cytogenet Genome Res. 2005;110(1-4):383-91. doi: 10.1159/000084970. Cytogenet Genome Res. 2005. PMID: 16093690 Review.
-
Evolution of R1 and R2 in the rDNA units of the genus Drosophila.Genetica. 1997;100(1-3):49-61. Genetica. 1997. PMID: 9440258 Review.
Cited by
-
Phylogeny of the Genus Drosophila.Genetics. 2018 May;209(1):1-25. doi: 10.1534/genetics.117.300583. Genetics. 2018. PMID: 29716983 Free PMC article. Review.
-
Unique transposon landscapes are pervasive across Drosophila melanogaster genomes.Nucleic Acids Res. 2015 Dec 15;43(22):10655-72. doi: 10.1093/nar/gkv1193. Epub 2015 Nov 17. Nucleic Acids Res. 2015. PMID: 26578579 Free PMC article.
-
Nacα protects the larval fat body from cell death by maintaining cellular proteostasis in Drosophila.Nat Commun. 2023 Sep 1;14(1):5328. doi: 10.1038/s41467-023-41103-1. Nat Commun. 2023. PMID: 37658058 Free PMC article.
-
Gene duplication in the major insecticide target site, Rdl, in Drosophila melanogaster.Proc Natl Acad Sci U S A. 2013 Sep 3;110(36):14705-10. doi: 10.1073/pnas.1311341110. Epub 2013 Aug 19. Proc Natl Acad Sci U S A. 2013. PMID: 23959864 Free PMC article.
-
Diversification and collapse of a telomere elongation mechanism.Genome Res. 2019 Jun;29(6):920-931. doi: 10.1101/gr.245001.118. Epub 2019 May 28. Genome Res. 2019. PMID: 31138619 Free PMC article.
References
-
- Craig NL, Craigie R, Gellert M, Lambowitz AM, Eds. Mobile DNA II. Washington: ASM Press; 2002.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

