The degree of microbiome complexity influences the epithelial response to infection
- PMID: 19689803
- PMCID: PMC2736203
- DOI: 10.1186/1471-2164-10-380
The degree of microbiome complexity influences the epithelial response to infection
Abstract
Background: The human microflora is known to be extremely complex, yet most pathogenesis research is conducted in mono-species models of infection. Consequently, it remains unclear whether the level of complexity of a host's indigenous flora can affect the virulence potential of pathogenic species. Furthermore, it remains unclear whether the colonization by commensal species affects a host cell's response to pathogenic species beyond the direct physical saturation of surface receptors, the sequestration of nutrients, the modulation of the physico-chemical environment in the oral cavity, or the production of bacteriocins. Using oral epithelial cells as a model, we hypothesized that the virulence of pathogenic species may vary depending on the complexity of the flora that interacts with host cells.
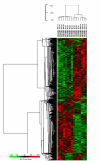
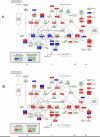
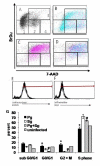
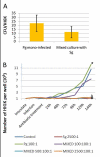
Results: This is the first report that determines the global epithelial transcriptional response to co-culture with defined complex microbiota. In our model, human immortalized gingival keratinocytes (HIGK) were infected with mono- and mixed cultures of commensal and pathogenic species. The global transcriptional response of infected cells was validated and confirmed phenotypically. In our model, commensal species were able to modulate the expression of host genes with a broad diversity of physiological functions and antagonize the effect of pathogenic species at the cellular level. Unexpectedly, the inhibitory effect of commensal species was not correlated with its ability to inhibit adhesion or invasion by pathogenic species.
Conclusion: Studying the global transcriptome of epithelial cells to single and complex microbial challenges offers clues towards a better understanding of how bacteria-bacteria interactions and bacteria-host interactions impact the overall host response. This work provides evidence that the degree of complexity of a mixed microbiota does influence the transcriptional response to infection of host epithelial cells, and challenges the current dogma regarding the potential versus the actual pathogenicity of bacterial species. These findings support the concept that members of the commensal oral flora have evolved cellular mechanisms that directly modulate the host cell's response to pathogenic species and dampen their relative pathogenicity.
Figures
Similar articles
-
Porphyromonas gingivalis initiates a mesenchymal-like transition through ZEB1 in gingival epithelial cells.Cell Microbiol. 2016 Jun;18(6):844-58. doi: 10.1111/cmi.12554. Epub 2016 Jan 13. Cell Microbiol. 2016. PMID: 26639759 Free PMC article.
-
Distinct gene expression characteristics in epithelial cell-Porphyromonas gingivalis interactions by integrating transcriptome analyses.Int J Med Sci. 2019 Sep 7;16(10):1320-1327. doi: 10.7150/ijms.33728. eCollection 2019. Int J Med Sci. 2019. PMID: 31692996 Free PMC article.
-
Distinct transcriptional profiles characterize oral epithelium-microbiota interactions.Cell Microbiol. 2005 Jun;7(6):811-23. doi: 10.1111/j.1462-5822.2005.00513.x. Cell Microbiol. 2005. PMID: 15888084
-
Beyond good and evil in the oral cavity: insights into host-microbe relationships derived from transcriptional profiling of gingival cells.J Dent Res. 2008 Mar;87(3):203-23. doi: 10.1177/154405910808700302. J Dent Res. 2008. PMID: 18296603 Free PMC article. Review.
-
Molecular interaction of Porphyromonas gingivalis with host cells: implication for the microbial pathogenesis of periodontal disease.J Periodontol. 2003 Jan;74(1):90-6. doi: 10.1902/jop.2003.74.1.90. J Periodontol. 2003. PMID: 12593602 Review.
Cited by
-
Spent culture supernatant of Streptococcus gordonii mitigates inflammation of human periodontal cells and inhibits proliferation of pathogenic oral microbes.J Periodontol. 2023 Apr;94(4):575-585. doi: 10.1002/JPER.22-0333. Epub 2023 Jan 6. J Periodontol. 2023. PMID: 36369979 Free PMC article.
-
Perturbation of the indigenous rat oral microbiome by ciprofloxacin dosing.Mol Oral Microbiol. 2013 Oct;28(5):404-14. doi: 10.1111/omi.12033. Epub 2013 Jul 12. Mol Oral Microbiol. 2013. PMID: 23844936 Free PMC article.
-
Cuticular bacteria appear detrimental to social spiders in mixed but not monoculture exposure.Curr Zool. 2016 Aug;62(4):377-384. doi: 10.1093/cz/zow015. Epub 2016 Mar 17. Curr Zool. 2016. PMID: 29491926 Free PMC article.
-
Comparing the microbiota of the cystic fibrosis lung and human gut.Gut Microbes. 2010 Mar;1(2):85-93. doi: 10.4161/gmic.1.2.11350. Epub 2010 Jan 29. Gut Microbes. 2010. PMID: 21326915 Free PMC article. No abstract available.
-
In Vitro Effects of Streptococcus oralis Biofilm on Peri-Implant Soft Tissue Cells.Cells. 2020 May 15;9(5):1226. doi: 10.3390/cells9051226. Cells. 2020. PMID: 32429151 Free PMC article.
References
-
- Krisanaprakornkit S, Kimball JR, Weinberg A, Darveau RP, Bainbridge BW, Dale BA. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect Immun. 2000;68:2907–2915. doi: 10.1128/IAI.68.5.2907-2915.2000. - DOI - PMC - PubMed
-
- Cosseau C, Devine DA, Dullaghan E, Gardy JL, Chikatamarla A, Gellatly S, Yu LL, Pistolic J, Falsafi R, Tagg J, Hancock RE. The commensal Streptococcus salivarius K12 downregulates the innate immune responses of human epithelial cells and promotes host-microbe homeostasis. Infect Immun. 2008;76:4163–4175. doi: 10.1128/IAI.00188-08. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

