MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli
- PMID: 19690171
- PMCID: PMC2781420
- DOI: 10.1074/jbc.M109.032904
MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli
Abstract
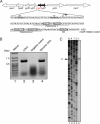
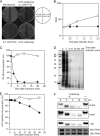
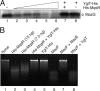
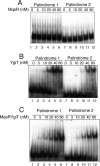
The mqsR gene has been shown to be positively regulated by the quorum-sensing autoinducer AI-2, which in turn activates a two-component system, the qseB-qseC operon. This operon plays an important role in biofilm formation in Escherichia coli. However, its cellular function has remained unknown. Here, we found that 1 base downstream of mqsR there is a gene, ygiT, that is co-transcribed with mqsR. Induction of mqsR caused cell growth arrest, whereas ygiT co-induction recovered cell growth. We demonstrate that MqsR (98 amino acid residues), which has no homology to the well characterized mRNA interferase MazF, is a potent inhibitor of protein synthesis that functions by degrading cellular mRNAs. In vivo and in vitro primer extension experiments showed that MqsR is an mRNA interferase specifically cleaving mRNAs at GCU. The mRNA interferase activity of purified MqsR was inhibited by purified YgiT (131 residues). MqsR forms a stable 2:1 complex with YgiT, and the complex likely functions as a repressor for the mqsR-ygiT operon by specifically binding to two different palindromic sequences present in the 5'-untranslated region of this operon.
Figures

References
-
- Davies D. G., Parsek M. R., Pearson J. P., Iglewski B. H., Costerton J. W., Greenberg E. P. (1998) Science 280, 295–298 - PubMed
-
- Hammer B. K., Bassler B. L. (2003) Mol. Microbiol. 50, 101–104 - PubMed
-
- Ren D., Bedzyk L. A., Thomas S. M., Ye R. W., Wood T. K. (2004) Appl. Microbiol. Biotechnol. 64, 515–524 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

