Binding of pro-prion to filamin A disrupts cytoskeleton and correlates with poor prognosis in pancreatic cancer
- PMID: 19690385
- PMCID: PMC2735930
- DOI: 10.1172/JCI39542
Binding of pro-prion to filamin A disrupts cytoskeleton and correlates with poor prognosis in pancreatic cancer
Abstract
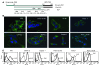
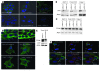
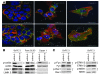
The cellular prion protein (PrP) is a highly conserved, widely expressed, glycosylphosphatidylinositol-anchored (GPI-anchored) cell surface glycoprotein. Since its discovery, most studies on PrP have focused on its role in neurodegenerative prion diseases, whereas its function outside the nervous system remains unclear. Here, we report that human pancreatic ductal adenocarcinoma (PDAC) cell lines expressed PrP. However, the PrP was neither glycosylated nor GPI-anchored, existing as pro-PrP and retaining its GPI anchor peptide signal sequence (GPI-PSS). We also showed that the PrP GPI-PSS has a filamin A-binding (FLNa-binding) motif and interacted with FLNa, an actin-associated protein that integrates cell mechanics and signaling. Binding of pro-PrP to FLNa disrupted cytoskeletal organization. Inhibition of PrP expression by shRNA in the PDAC cell lines altered the cytoskeleton and expression of multiple signaling proteins; it also reduced cellular proliferation and invasiveness in vitro as well as tumor growth in vivo. A subgroup of human patients with pancreatic cancer was found to have tumors that expressed pro-PrP. Most importantly, PrP expression in tumors correlated with a marked decrease in patient survival. We propose that binding of pro-PrP to FLNa perturbs FLNa function, thus contributing to the aggressiveness of PDAC. Prevention of this interaction could provide an attractive target for therapeutic intervention in human PDAC.
Figures





Similar articles
-
Binding of pro-prion to filamin A: by design or an unfortunate blunder.Oncogene. 2010 Sep 30;29(39):5329-45. doi: 10.1038/onc.2010.307. Epub 2010 Aug 9. Oncogene. 2010. PMID: 20697352 Free PMC article. Review.
-
The fatal attraction between pro-prion and filamin A: prion as a marker in human cancers.Biomark Med. 2010 Jun;4(3):453-64. doi: 10.2217/bmm.10.14. Biomark Med. 2010. PMID: 20550479 Free PMC article.
-
Pro-prion binds filamin A, facilitating its interaction with integrin beta1, and contributes to melanomagenesis.J Biol Chem. 2010 Sep 24;285(39):30328-39. doi: 10.1074/jbc.M110.147413. Epub 2010 Jul 21. J Biol Chem. 2010. PMID: 20650901 Free PMC article.
-
Glycosylphosphatidylinositol Anchor Modification Machinery Deficiency Is Responsible for the Formation of Pro-Prion Protein (PrP) in BxPC-3 Protein and Increases Cancer Cell Motility.J Biol Chem. 2016 Feb 19;291(8):3905-17. doi: 10.1074/jbc.M115.705830. Epub 2015 Dec 18. J Biol Chem. 2016. PMID: 26683373 Free PMC article.
-
Prion Protein Family Contributes to Tumorigenesis via Multiple Pathways.Adv Exp Med Biol. 2017;1018:207-224. doi: 10.1007/978-981-10-5765-6_13. Adv Exp Med Biol. 2017. PMID: 29052140 Review.
Cited by
-
The Cellular Prion Protein and the Hallmarks of Cancer.Cancers (Basel). 2021 Oct 8;13(19):5032. doi: 10.3390/cancers13195032. Cancers (Basel). 2021. PMID: 34638517 Free PMC article. Review.
-
Disruption of prion protein-HOP engagement impairs glioblastoma growth and cognitive decline and improves overall survival.Oncogene. 2015 Jun;34(25):3305-14. doi: 10.1038/onc.2014.261. Epub 2014 Aug 25. Oncogene. 2015. PMID: 25151961
-
CRABP-II enhances pancreatic cancer cell migration and invasion by stabilizing interleukin 8 expression.Oncotarget. 2016 Dec 26;8(32):52432-52444. doi: 10.18632/oncotarget.14194. eCollection 2017 Aug 8. Oncotarget. 2016. PMID: 28881741 Free PMC article.
-
Prion protein scrapie and the normal cellular prion protein.Prion. 2016;10(1):63-82. doi: 10.1080/19336896.2015.1110293. Prion. 2016. PMID: 26645475 Free PMC article. Review.
-
High Expression of PRNP Predicts Poor Prognosis in Korean Patients with Gastric Cancer.Cancers (Basel). 2022 Jun 28;14(13):3173. doi: 10.3390/cancers14133173. Cancers (Basel). 2022. PMID: 35804944 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

