Massively parallel resequencing of the isogenic Drosophila melanogaster strain w(1118); iso-2; iso-3 identifies hotspots for mutations in sensory perception genes
- PMID: 19690466
- PMCID: PMC3839425
- DOI: 10.4161/fly.3.3.9652
Massively parallel resequencing of the isogenic Drosophila melanogaster strain w(1118); iso-2; iso-3 identifies hotspots for mutations in sensory perception genes
Abstract
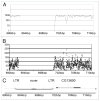
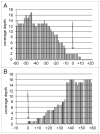
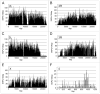
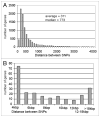
We used the Illumina reversible-short sequencing technology to obtain 17-fold average depth (s.d. approximately 8) of approximately 94% of the euchromatic genome and approximately 1-5% of the heterochromatin sequence of the Drosophila melanogaster isogenic strain w(1118); iso-2; iso-3. We show that this strain has a approximately 9 kb deletion that uncovers the first exon of the white (w) gene, approximately 4 kb of downstream promoter sequences, and most of the first intron, thus demonstrating that whole-genome sequencing can be used for mutation characterization. We chose this strain because there are thousands of transposon insertion lines and hundreds of isogenic deficiency lines available with this genetic background, such as the Exelixis, Inc., and the DrosDEL collections. We compared our sequence to Release 5 of the finished reference genome sequence which was made from the isogenic strain y(1); cn(1) bw(1) sp(1) and identified 356,614 candidate SNPs in the approximately 117 Mb unique sequence genome, which represents a substitution rate of approximately 1/305 nucleotides ( approximately 0.30%). The distribution of SNPs is not uniform, but rather there is a approximately 2-fold increase in SNPs on the autosome arms compared with the X chromosome and a approximately 7-fold increase when compared to the small 4(th) chromosome. This is consistent with previous analyses that demonstrated a correlation between recombination frequency and SNP frequency. An unexpected finding was a SNP hotpot in a approximately 20 Mb central region of the 4(th) chromosome, which might indicate higher than expected recombination frequency in this region of this chromosome. Interestingly, genes involved in sensory perception are enriched in SNP hotspots and genes encoding developmental genes are enriched in SNP coldspots, which suggests that recombination frequencies might be proportional to the evolutionary selection coefficient. There are currently 12 Drosophila species sequenced, and this represents one of many isogenic Drosophila melanogaster genome sequences that are in progress. Because of the dramatic increase in power in using isogenic lines rather than outbred individuals, the SNP information should be valuable as a test bed for understanding genotype-by-environment interactions in human population studies.
Figures



References
-
- International Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 431:931–45. - PubMed
-
- Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–204. - PubMed
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–95. - PubMed
-
- Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
