HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins
- PMID: 19690614
- PMCID: PMC2723923
- DOI: 10.1371/journal.pone.0006687
HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins
Abstract
Background: Despite the extensive genetic diversity of HIV-1, viral evolution in response to immune selective pressures follows broadly predictable mutational patterns. Sites and pathways of Human Leukocyte-Antigen (HLA)-associated polymorphisms in HIV-1 have been identified through the analysis of population-level data, but the full extent of immune escape pathways remains incompletely characterized. Here, in the largest analysis of HIV-1 subtype B sequences undertaken to date, we identify HLA-associated polymorphisms in the three HIV-1 proteins most commonly considered in cellular-based vaccine strategies. Results are organized into protein-wide escape maps illustrating the sites and pathways of HLA-driven viral evolution.
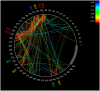
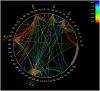
Methodology/principal findings: HLA-associated polymorphisms were identified in HIV-1 Gag, Pol and Nef in a multicenter cohort of >1500 chronically subtype-B infected, treatment-naïve individuals from established cohorts in Canada, the USA and Western Australia. At q< or =0.05, 282 codons commonly mutating under HLA-associated immune pressures were identified in these three proteins. The greatest density of associations was observed in Nef (where close to 40% of codons exhibited a significant HLA association), followed by Gag then Pol (where approximately 15-20% of codons exhibited HLA associations), confirming the extensive impact of immune selection on HIV evolution and diversity. Analysis of HIV codon covariation patterns identified over 2000 codon-codon interactions at q< or =0.05, illustrating the dense and complex networks of linked escape and secondary/compensatory mutations.
Conclusions/significance: The immune escape maps and associated data are intended to serve as a user-friendly guide to the locations of common escape mutations and covarying codons in HIV-1 subtype B, and as a resource facilitating the systematic identification and classification of immune escape mutations. These resources should facilitate research in HIV epitope discovery and host-pathogen co-evolution, and are relevant to the continued search for an effective CTL-based AIDS vaccine.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- 1U01-AI069484/AI/NIAID NIH HHS/United States
- AI27661/AI/NIAID NIH HHS/United States
- U01 AI046376/AI/NIAID NIH HHS/United States
- U01 AI069477/AI/NIAID NIH HHS/United States
- U01 AI069502/AI/NIAID NIH HHS/United States
- U01 AI069513/AI/NIAID NIH HHS/United States
- AI 69467-01/AI/NIAID NIH HHS/United States
- UM1 AI069494/AI/NIAID NIH HHS/United States
- UM1 AI069423/AI/NIAID NIH HHS/United States
- AI38858-09S1/AI/NIAID NIH HHS/United States
- U01 AI069474/AI/NIAID NIH HHS/United States
- AI32782/AI/NIAID NIH HHS/United States
- U01 AI069434/AI/NIAID NIH HHS/United States
- AI-69465/AI/NIAID NIH HHS/United States
- AI069556/AI/NIAID NIH HHS/United States
- U01 AI069447/AI/NIAID NIH HHS/United States
- UM1 AI069501/AI/NIAID NIH HHS/United States
- UM1 AI069472/AI/NIAID NIH HHS/United States
- U01 AI069467/AI/NIAID NIH HHS/United States
- U01 AI069423/AI/NIAID NIH HHS/United States
- UM1 AI069513/AI/NIAID NIH HHS/United States
- M01 RR000096/RR/NCRR NIH HHS/United States
- M01 RR000046/RR/NCRR NIH HHS/United States
- AI46370/AI/NIAID NIH HHS/United States
- U01 AI027661/AI/NIAID NIH HHS/United States
- UM1 AI069424/AI/NIAID NIH HHS/United States
- AI69472/AI/NIAID NIH HHS/United States
- AI-069439/AI/NIAID NIH HHS/United States
- U01 AI069465/AI/NIAID NIH HHS/United States
- UM1 AI069434/AI/NIAID NIH HHS/United States
- UL1 RR024996/RR/NCRR NIH HHS/United States
- K24 AI064086/AI/NIAID NIH HHS/United States
- UM1 AI069411/AI/NIAID NIH HHS/United States
- 1 U01 AI069452-01/AI/NIAID NIH HHS/United States
- UM1 AI069432/AI/NIAID NIH HHS/United States
- AI069532/AI/NIAID NIH HHS/United States
- AI069434/AI/NIAID NIH HHS/United States
- AI046376-05/AI/NIAID NIH HHS/United States
- AI 068634/AI/NIAID NIH HHS/United States
- M01 RR-00032/RR/NCRR NIH HHS/United States
- AI069474/AI/NIAID NIH HHS/United States
- UM1 AI069495/AI/NIAID NIH HHS/United States
- U01 AI069484/AI/NIAID NIH HHS/United States
- AI36214/AI/NIAID NIH HHS/United States
- UM1 AI069471/AI/NIAID NIH HHS/United States
- U01 AI069439/AI/NIAID NIH HHS/United States
- U01 AI069556/AI/NIAID NIH HHS/United States
- M01-RR00096/RR/NCRR NIH HHS/United States
- RR00051/RR/NCRR NIH HHS/United States
- AI 069501/AI/NIAID NIH HHS/United States
- U01 AI069532/AI/NIAID NIH HHS/United States
- UM1 AI069439/AI/NIAID NIH HHS/United States
- K24-AI 064086/AI/NIAID NIH HHS/United States
- M01 RR002635/RR/NCRR NIH HHS/United States
- P30 AI045008/AI/NIAID NIH HHS/United States
- M01 RR000044/RR/NCRR NIH HHS/United States
- UM1 AI068634/AI/NIAID NIH HHS/United States
- U01 AI069501/AI/NIAID NIH HHS/United States
- AI69411/AI/NIAID NIH HHS/United States
- AI34853/AI/NIAID NIH HHS/United States
- M01 RR000750/RR/NCRR NIH HHS/United States
- AI069495/AI/NIAID NIH HHS/United States
- UM1 AI069477/AI/NIAID NIH HHS/United States
- U01 AI069432/AI/NIAID NIH HHS/United States
- AI068636/AI/NIAID NIH HHS/United States
- U01 AI069450/AI/NIAID NIH HHS/United States
- U01 AI038858/AI/NIAID NIH HHS/United States
- RR024975/RR/NCRR NIH HHS/United States
- AI69494-01/AI/NIAID NIH HHS/United States
- RR02635/RR/NCRR NIH HHS/United States
- U01 AI046370/AI/NIAID NIH HHS/United States
- U01 AI068636/AI/NIAID NIH HHS/United States
- UM1 AI069474/AI/NIAID NIH HHS/United States
- AI060354/AI/NIAID NIH HHS/United States
- M01 RR000052/RR/NCRR NIH HHS/United States
- U01 AI034853/AI/NIAID NIH HHS/United States
- AI069419-01/AI/NIAID NIH HHS/United States
- AI069424/AI/NIAID NIH HHS/United States
- R01 AI060460/AI/NIAID NIH HHS/United States
- M01 RR000051/RR/NCRR NIH HHS/United States
- U01 AI069495/AI/NIAID NIH HHS/United States
- UM1 AI069556/AI/NIAID NIH HHS/United States
- P30 AI110527/AI/NIAID NIH HHS/United States
- M01 RR000032/RR/NCRR NIH HHS/United States
- U01 AI027673/AI/NIAID NIH HHS/United States
- AI069502-01/AI/NIAID NIH HHS/United States
- 5-M01 RR00044/RR/NCRR NIH HHS/United States
- P30 AI036214/AI/NIAID NIH HHS/United States
- U0I AI38858/AI/NIAID NIH HHS/United States
- UM1 AI069502/AI/NIAID NIH HHS/United States
- U01 AI025859/AI/NIAID NIH HHS/United States
- UM1 AI069450/AI/NIAID NIH HHS/United States
- UM1 AI069532/AI/NIAID NIH HHS/United States
- AI27673/AI/NIAID NIH HHS/United States
- UM1 AI069465/AI/NIAID NIH HHS/United States
- U01 AI069424/AI/NIAID NIH HHS/United States
- U01 AI032782/AI/NIAID NIH HHS/United States
- UL1 RR024975/RR/NCRR NIH HHS/United States
- AI-069513/AI/NIAID NIH HHS/United States
- AI069450/AI/NIAID NIH HHS/United States
- RR-00052/RR/NCRR NIH HHS/United States
- U01 AI069411/AI/NIAID NIH HHS/United States
- AI069472/AI/NIAID NIH HHS/United States
- 5-P30-AI-045008-07/AI/NIAID NIH HHS/United States
- UM1 AI069419/AI/NIAID NIH HHS/United States
- AI069432/AI/NIAID NIH HHS/United States
- AI50410/AI/NIAID NIH HHS/United States
- P30 AI060354/AI/NIAID NIH HHS/United States
- U01 AI069419/AI/NIAID NIH HHS/United States
- R01 AI030914/AI/NIAID NIH HHS/United States
- AI25859/AI/NIAID NIH HHS/United States
- U01 AI069494/AI/NIAID NIH HHS/United States
- UM1 AI069447/AI/NIAID NIH HHS/United States
- U01 AI069452/AI/NIAID NIH HHS/United States
- UM1 AI069467/AI/NIAID NIH HHS/United States
- AI69423-01/AI/NIAID NIH HHS/United States
- AI069447-01/AI/NIAID NIH HHS/United States
- AI069477/AI/NIAID NIH HHS/United States
- RR024996/RR/NCRR NIH HHS/United States
- RR00046/RR/NCRR NIH HHS/United States
- AI069471/AI/NIAID NIH HHS/United States
- U01 AI069471/AI/NIAID NIH HHS/United States
- U01 AI069472/AI/NIAID NIH HHS/United States
- P30 AI050410/AI/NIAID NIH HHS/United States
- U01 AI068634/AI/NIAID NIH HHS/United States
- UM1 AI068636/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

