Longicorn beetle that vectors pinewood nematode carries many Wolbachia genes on an autosome
- PMID: 19692404
- PMCID: PMC2817283
- DOI: 10.1098/rspb.2009.1022
Longicorn beetle that vectors pinewood nematode carries many Wolbachia genes on an autosome
Abstract
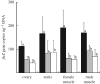
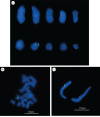
Monochamus alternatus is the longicorn beetle notorious as a vector of the pinewood nematode that causes the pine wilt disease. When two populations of M. alternatus were subjected to diagnostic polymerase chain reaction (PCR) detection of four Wolbachia genes, only the ftsZ gene was detected from one of the populations. The Wolbachia ftsZ gene persisted even after larvae were fed with a tetracycline-containing diet for six weeks. The inheritance of the ftsZ gene was not maternal but biparental, exhibiting a typical Mendelian pattern. The ftsZ gene titres in homozygotic ftsZ(+) insects were nearly twice as high as those in heterozygotic ftsZ(+) insects. Exhaustive PCR surveys revealed that 31 and 30 of 214 Wolbachia genes examined were detected from the two insect populations, respectively. Many of these Wolbachia genes contained stop codon(s) and/or frame shift(s). Fluorescent in situ hybridization confirmed the location of the Wolbachia genes on an autosome. On the basis of these results, we conclude that a large Wolbachia genomic region has been transferred to and located on an autosome of M. alternatus. The discovery of massive gene transfer from Wolbachia to M. alternatus would provide further insights into the evolution and fate of laterally transferred endosymbiont genes in multicellular host organisms.
Figures



References
-
- Abascal F., Zardoya R., Posada D.2005ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 (doi:10.1093/bioinformatics/bti263) - DOI - PubMed
-
- Andersson J. O.2005Lateral gene transfer in eukaryotes. Cell. Mol. Life Sci. 62, 1182–1197 (doi:10.1007/s00018-005-4539-z) - DOI - PMC - PubMed
-
- Beard C. B., O'Neill S. L., Tesh R. B., Richards F. F., Aksoy S.1993Modification of arthropod vector competence via symbiotic bacteria. Parasitol. Today 9, 179–183 (doi:10.1016/0169-4758(93)90142-3) - DOI - PubMed
-
- Bourtzis K., Miller T. A.2003Insect symbiosis Boca Raton, FL: CRC Press
-
- Dobson S. L.2003Reversing Wolbachia-based population replacement. Trends Parasitol 19, 128–133 (doi:10.1016/S1471-4922(03)00002-3) - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
