Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs
- PMID: 19693008
- PMCID: PMC2746258
- DOI: 10.1038/nature08321
Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs
Abstract
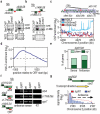
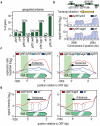
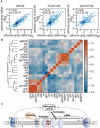
Eukaryotic transcriptomes are characterized by widespread transcription of noncoding and antisense RNAs, which is linked to key chromosomal processes, such as chromatin remodelling, gene regulation and heterochromatin assembly. However, these transcripts can be deleterious, and their accumulation is suppressed by several mechanisms including degradation by the nuclear exosome. The mechanisms by which cells differentiate coding RNAs from transcripts targeted for degradation are not clear. Here we show that the variant histone H2A.Z, which is loaded preferentially at the 5' ends of genes by the Swr1 complex containing a JmjC domain protein, mediates suppression of antisense transcripts in the fission yeast Schizosaccharomyces pombe genome. H2A.Z is partially redundant in this regard with the Clr4 (known as SUV39H in mammals)-containing heterochromatin silencing complex that is also distributed at euchromatic loci, and with RNA interference component Argonaute (Ago1). Loss of Clr4 or Ago1 alone has little effect on antisense transcript levels, but cells lacking either of these factors and H2A.Z show markedly increased levels of antisense RNAs that are normally degraded by the exosome. These analyses suggest that as well as performing other functions, H2A.Z is a component of a genome indexing mechanism that cooperates with heterochromatin and RNAi factors to suppress read-through antisense transcripts.
Figures

References
-
- Wilhelm BT, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. - PubMed
-
- Cawley S, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. - PubMed
-
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. - PubMed
-
- Hirota K, et al. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature. 2008;456:130–134. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

